In Situ Sequencing
The in situ sequencing (ISS) unit provides spatially resolved gene expression data for panels of genes at subcellular resolution. The technique has been developed in the lab of Mats Nilsson who has pioneered the field of generating in situ gene expression and mutation profiles (Ke R et al; 2013 Nature Methods).
Unique features of ISS
- Preservation of spatial information in tissue context
- Targeted approach, using padlock probing
- Highly specific amplification tool, offering multiple levels of molecular specificity
- Multiplexing, up to few hundred transcripts per sample
- Sensitivity, tunable
- High throughput due to wide-field imaging
In situ sequencing enables localization and quantification of more than 100 transcripts simultaneously with subcellular resolution in a single tissue section per experiment.
Technology description
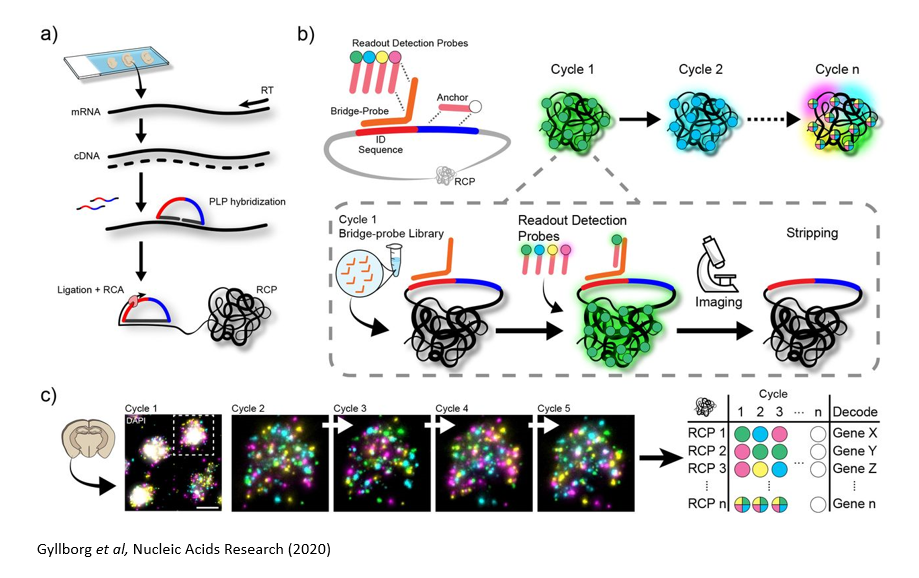
a | Sample pre-treatment & library preparation
Tissue/cell line samples on standard microscopy slides are pre-treated to generate cDNA in situ. A set of genes are targeted with a custom designed padlock probe library. Probes that have specifically interacted with the targeted transcripts are amplified by Rolling Circle Amplification (RCA) reaction, and the specific amplification generates signals with high signal to noise ratio.
b & c | In situ barcode sequencing reaction & Imaging
Specific barcode sequence in the probe for each targeted gene is decoded by sequential hybridization reaction and imaging cycles. The signals are detected by fluorescently labelled oligonucleotide libraries. ISS can be used to detect up to a few hundred genes per sample. Wide-field imaging enables high throughput.
Image analysis
After image acquisition, images are processed with the analysis pipeline. CSV file is generated for x-y coordinates map of each signal, and each cell nucleus.
Services
- Initial consultation and price quote (free)
- Discussion of project/experimental design (including a custom probe design service)
- In house library preparation training (generating RCA products in the tissues)
- Barcode-sequencing and imaging
- Mapping coordinates of gene expression in the tissue (primary image analysis)
- Add on services: H&E and IHC imaging
New projects from users are assessed with an initial sample screening and a feasibility test.
Applications
ISS technology has been successfully demonstrated for a variety of applications such as mapping of
- Molecularly defined cell types
- RNA editing
- Mutational heterogeneity
- Splice-variant
across different types of tissue sections including fresh frozen, PFA-fixed and FFPE.
Equipments
- Two automated epifluorescence microscopes (Axio imager.Z2; Zeiss and DMi8; Leica) equipped with multiwavelength LED light with 6 separate wavelengths.
- Automated slide stainer with a liquid handling system (Autostainer 360; LAB Vision) that can process 36 slides at one go.
- Two Xenium Analysers from 10X Genomics for fast, automated and high sensitivity in situ sequencing analysis
Questions?
Interested to know more about insitu sequencing technology? Please contact the unit using this email (iss@scilifelab.se). We will get back to you shortly to schedule a meeting for further discussion.
