Daniel Fürth
We study how information is stored, processed, and transmitted between cells with a special focus on how the aforementioned information processing enables computations in the nervous system.
To do this we develop novel tools and techniques at the intersection of molecular biology and computer science.
⚗️ Research 🔬
Understanding the nature of cell communication in health and disease remains one of the most important areas of biology.
Our lab searches for hitherto unknown mechanisms by which symbolic information can be transferred between cells.
Identification of such transmission would enable us to read and write those messages.
🛠️ Tools 🧰
The development of novel tools is an intricate part of our lab.
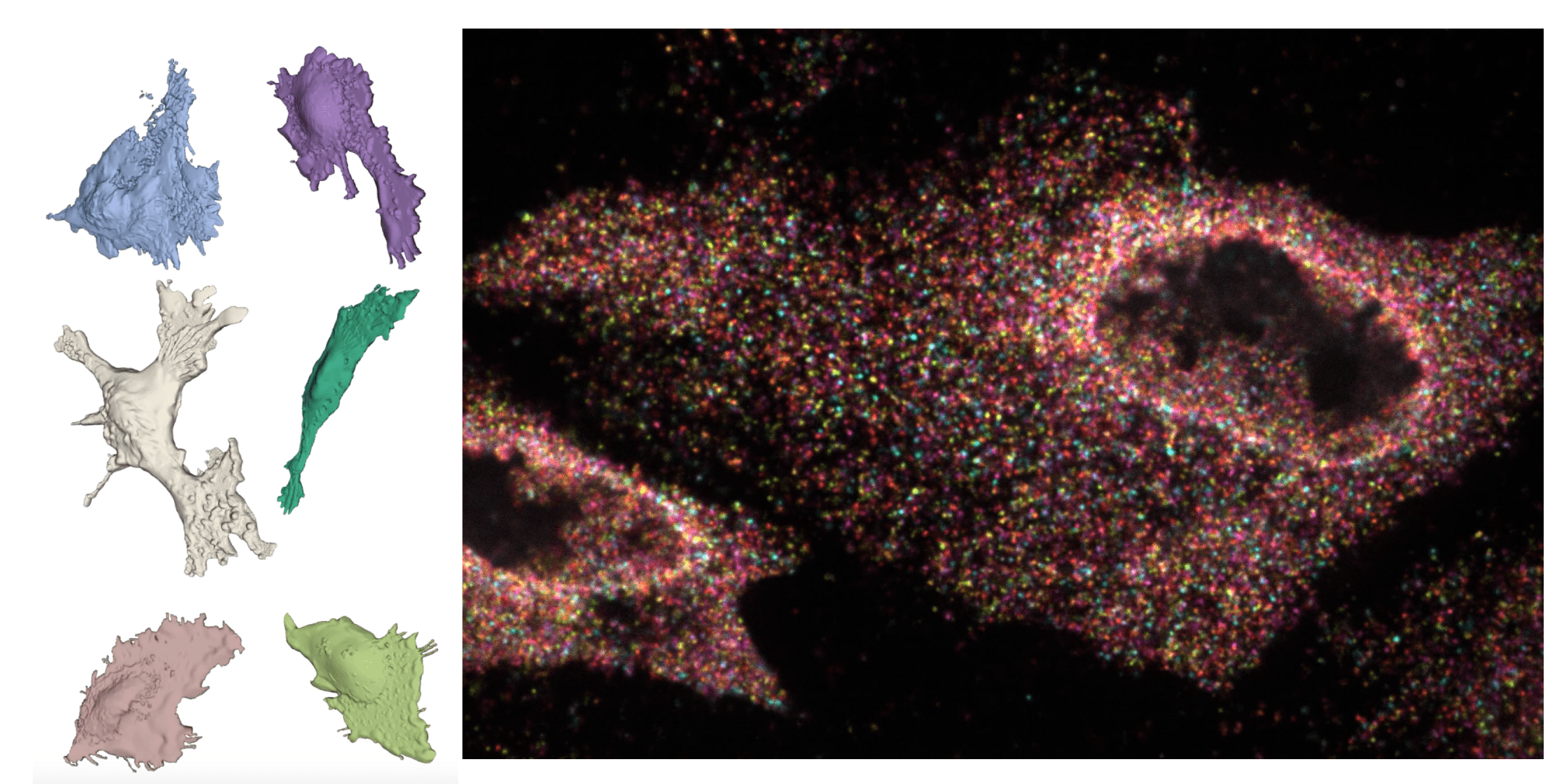
🔬 Single-molecule imaging
We use and develop methods to read and write information at the single molecule level.
🎇 Programmable bioconjugation
We combine nonlinear optical microscopy together with chemoselective moieties to develop new ways to perturb and interrogate information at the single-molecule level.
🧠 Computational anatomy
Whole-mount 3D single-molecule data at the transcriptome level necessitates new ways of compiling and analyzing anatomical data. We develop computational tools and resources.