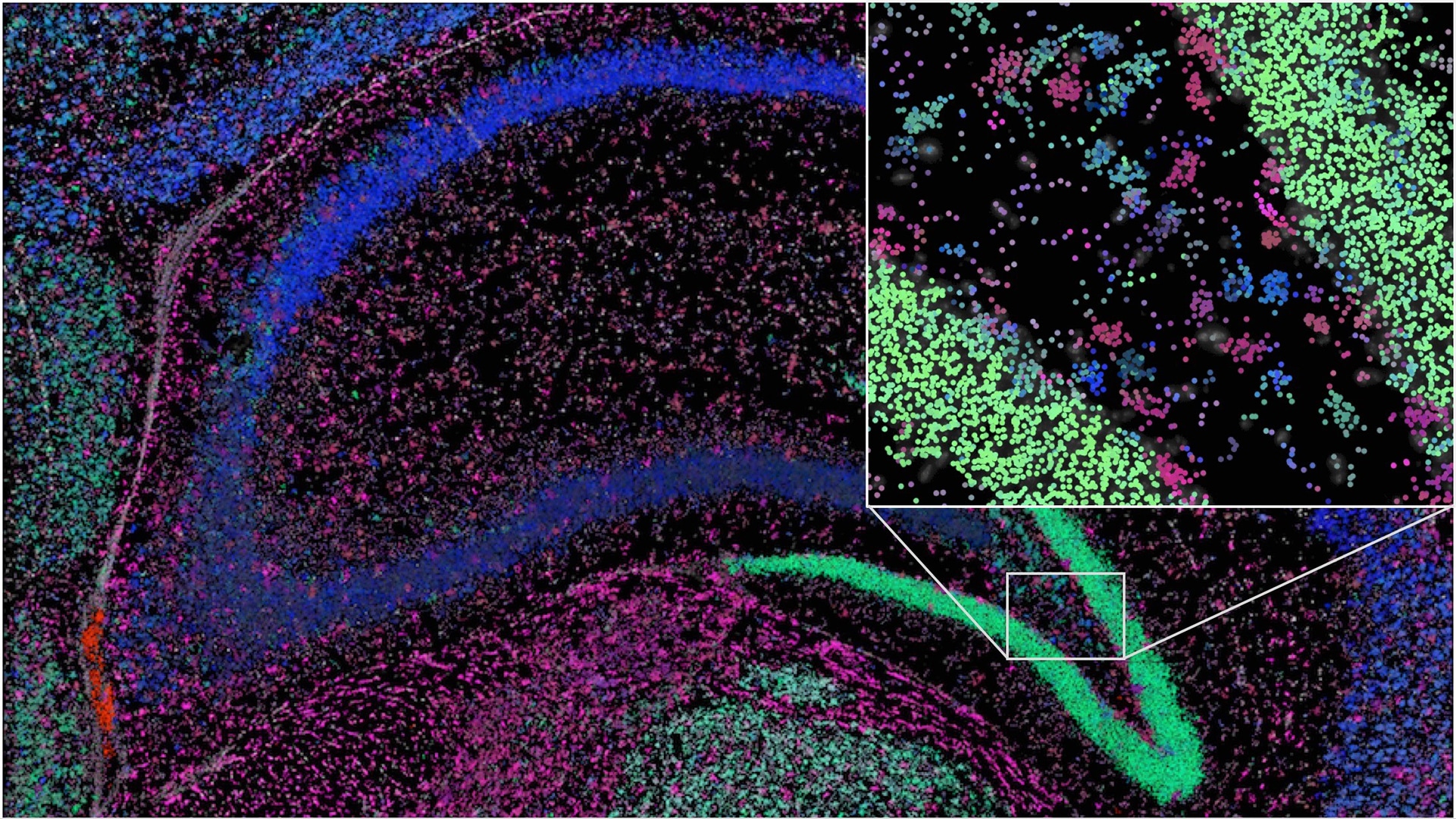
Enhanced social network AI method used for identifying cell types in situ
To facilitate the interpretation of the vast amounts of microscopy images containing gene expression data from a number of different tissue types, SciLifelab researchers have developed a new image analysis method. The new method is based on algorithms used in artificial intelligence and was originally devised to enhance understanding of social networks.
Our organs consist of trillions of cells constantly carrying out a large number of vital tasks. Gene expression occurs through mRNA transcription which is later translated into proteins in the cytoplasm. The combination of mRNAs expressed in every cell defines its function and identity.
In a recent study, published in The FEBS Journal, SciLifeLab researcher Carolina Wählby and Gabriele Partel (Uppsala University) investigated the possibility to further develop a microscopy method capable of showing millions of detected mRNA sequences at once. The problem with the existing method is that it is very hard to distinguish between all the important details in such a complex image. The researcher wanted to solve this problem by implementing AI-based detection capable of automatically detecting different cell types and even functions and interactions between individual cells.
“We’re using the latest AI methods – specifically, graph neural networks, developed to analyse social networks; and adapting them to understand biological patterns and successive variation in tissue samples. The cells are comparable to social groupings that can be defined according to the activities they share in their social networks like Twitter, sharing their Google search results or TV recommendations”, says Carolina Wählby, in a press release from Uppsala University.
Earlier AI-based analysis methods required knowledge about tissue cell types and cell nuclei had to be identified in advance. The conventionally used method, “single-cell analysis”, may lose some of the mRNA and fail to detect certain cell types. And even with advanced automated image analysis, nuclei can be hard to detect, especially if the cells are packed densely together.
“With our analysis, which we call ‘spage2vec’, we can now get corresponding results without any previous knowledge of expected cell types. And what’s more, we can find new cell types and intra- or intercellular functions in tissue”, Says Carolina Wählby.
The researchers are currently working on perfecting the new method by investigating differentiation and organization of various types of cells during the early development of the heart. They are also working in close collaboration with cancer researchers aiming to implement the method to better understand how tumour tissue interacts with surrounding healthy tissue, at molecular level. The aim is that, in the long term, this will lead to better treatments that can be adapted to individual patients.