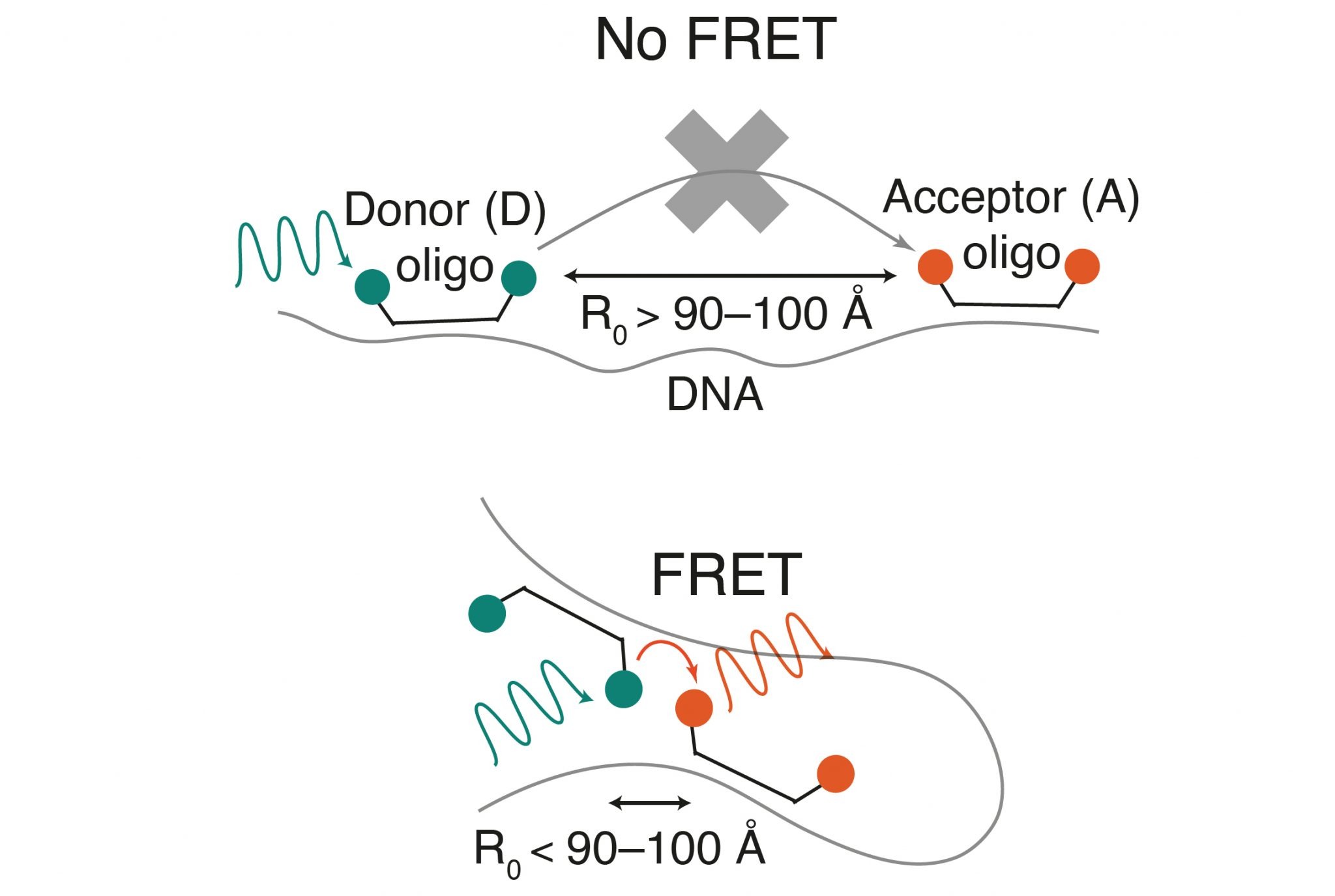
FRET-FISH measures targeted chromatin compaction in single cells
SciLifeLab researchers from Karolinska Institutet have developed a new method capable of measuring chromatin compaction at specific DNA loci in single cells. The method is based on a combination of the commonly used FISH (spatial DNA mapping) and FRET (fluorescence energy transfer detection) techniques and is called FRET-FISH.
Defined as the amount of DNA per volume in the nucleus, chromatin compaction is a fundamental genomic property that correlates with gene activity. Loosely packed genomic regions (euchromatin) usually contain many genes and are transcriptionally active, while regions containing highly compacted chromatin (heterochromatin) contain almost no genes and are transcriptionally silent.
To study chromatin compaction, DNA intercalating dyes, such as DAPI, can be used to visualize chromatin density under a fluorescence microscope. DNA transposases can also be used to insert specific DNA sequences in the more accessible chromatin regions, which can be followed by massively parallel sequencing of the targeted genomic regions.
DNA intercalating dyes are very effective for distinguishing less compact chromatin regions from more condensed ones, but they are not specific enough to target selected genomic locations. DNA transposase-based methods, such as Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), can measure DNA accessibility genome wide even in single cells, but they cannot probe chromatin compaction directly.
In order to combine these two techniques and measure chromatin compaction directly at selected genomic locations, researchers from Karolinska Institutet, led by SciLifeLab Fellow Alumna Magda Bienko and SciLifeLab researcher Nicola Crosetto, developed a new methodology, known as FRET-FISH, which combines DNA fluorescence in situ hybridization (FISH) with fluorescence energy transfer (FRET).
FISH is a common method, used to precisely map the spatial position of any DNA sequence on the genomic 3D structure inside the nucleus, while FRET has been extensively used to study protein-protein and protein-DNA interactions, as well as conformational changes in proteins and nucleic acids, by relying on energy transfer between a light-excited donor and an acceptor fluorophore. Up until the recent study, published in Nature Communications, a combination of the two methods has never been reported.
“FRET measurements are notoriously challenging in fixed cells and a combination of FRET and DNA FISH has not been described before” says co-last author, Magda Bienko.
To optimize the new method, first author Ana Mota (Karolinska Institutet) and her colleagues, began testing different probe designs with varying number of oligonucleotides coupled to FRET donor and acceptor dyes with different spacing distances between them. They managed to show that FRET-FISH can be used to detect local chromatin compaction changes upon treatment of cells with drugs that induce global changes in chromatin compaction, during different cell cycle phases, and as cells progressively age, thus assessing the reproducibility of the method, as well as validating it.
“Beyond studying local chromatin compaction, we envision that FRET-FISH could also be applied to study enhancer-promoter contacts or chromatin loop organization in single cells, without the need to rely on more sophisticated microscopy techniques” says co-last author Nicola Crosetto.
“I believe that FRET-FISH is a powerful tool that opens many new possibilities to study the biophysical properties of chromatin and understand how the three-dimensional structure of the genome influences its functions” concludes Magda Bienko.
The research was supported by grants from the Swedish Research Council, the Swedish Cancer Society, Science for Life Laboratory, Karolinska Institutet KID Funding Program, the Human Frontier Science Program, the Ragnar Söderberg Foundation, and the European Research Council under the European Union’s Horizon 2020 research and innovation program.