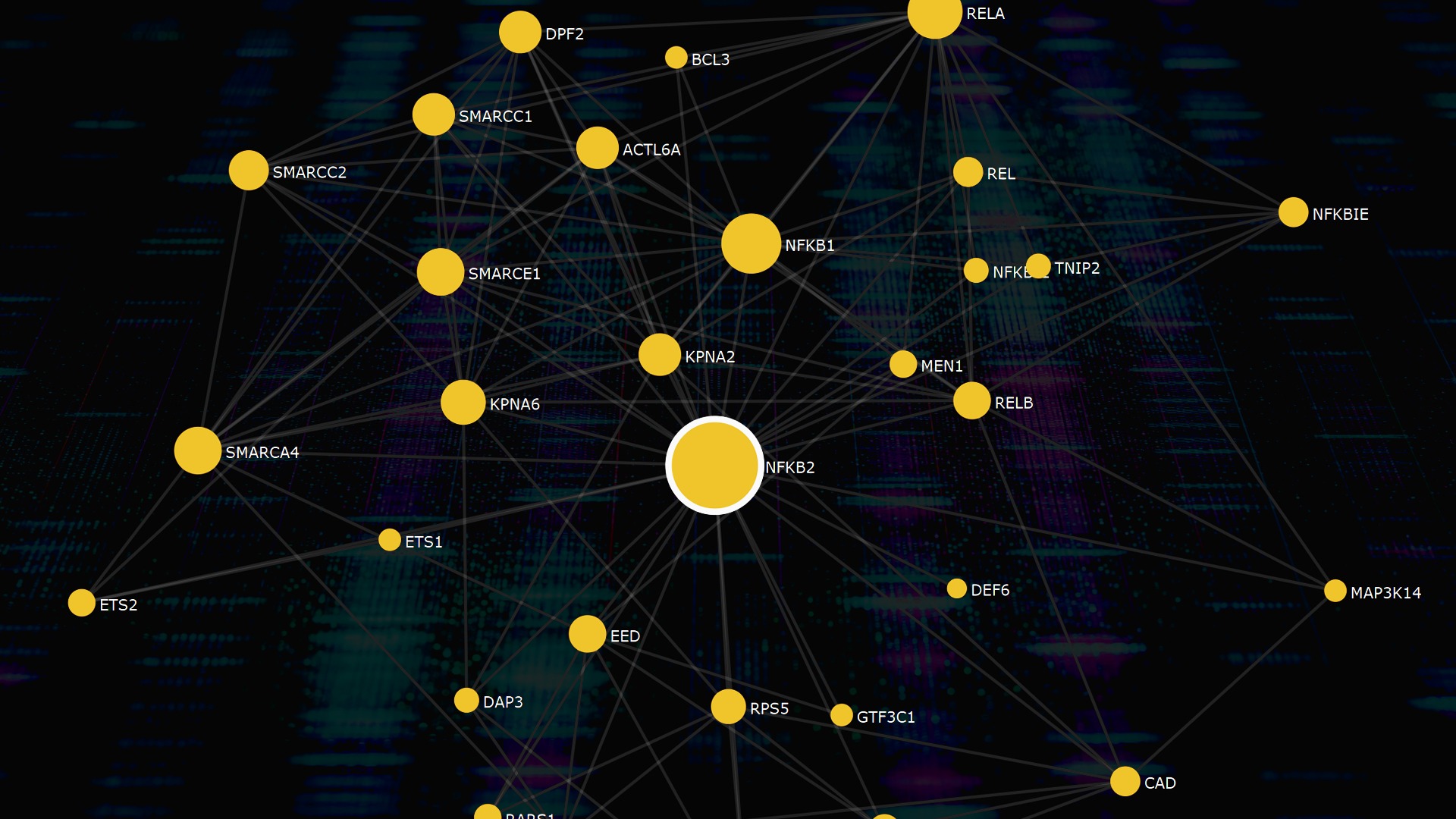
FunCoup 5 networks: New species, updated source data and improved functionality
SciLifeLab researcher Erik Sonnhammer (Stockholm University) and his team has released a new version of FunCoup, one of the most comprehensive functional association networks of genes/proteins available. The new version is described in the Journal of Molecular Biology.
The new version is called FunCoup 5 and includes four new species; two eukaryotes: Sus scrofa and Dictyostelium discoideum and two archaea: Methanocaldococcus jannaschii and Saccharolobus solfataricus, in addition to the already 17 existing species.
As an extension to the human interactome, FunCoup 5 also includes the SARS-CoV-2 proteome, allowing users to visualize and analyze interactions between the virus and human proteins in order to better understand COVID-19.
The availability of source data has improved drastically since the last database update and the demand for a functional association tool has grown among users. To meet these requirements, the researchers included updated source data for evidences, gold standards, and genomes as well as improved functionality to the new release of FunCoup.
In the new release, directed regulatory links inferred from transcription factor binding can be visualized in the network viewer for the human interactome, and for ten species the network can be filtered for genes expressed in a certain tissue. This new release of FunCoup constitutes a major advance for the users.