Lost and found in life science
This fall’s SciLifeLab Day offered a number of new concepts. Interactive workshops and four-minutes research presentations mingled with traditional talks on frontline research.
Over 500 participants attended the two-day event. SciLifeLab’s technologies and services – how to use them and how they should develop – were discussed in different formats. One service offered on site was the bioinformatic consultation group, which was available for discussions during breaks.
Some of SciLifeLab’s strategic recruitments took the chance to present their research to the community at Fellows corner. The challenge was to present their research as riveting, inspiring, and pedagogic as possible – in four minutes.
Carolina Wählby showed the sign for congestion tax and talked about how imaging and processing algorithms are used to register who should pay the tax. She compared that to her own research where she automates image analysis of cells. She counseled everyone to, the next time they see the sign, think about cells, new drugs and high throughput analysis. And also that these things are done at SciLifeLab.
Another fellow, Paul Hudson, talked about engineering cyanobacteria into making gasoline and encouraged students to “learn Python and Matlab, because math is definitely present in modern biology.”
Sophie Sanchez handed out 3d-glasses and showed how she interprets x-ray scans of fossils.
If you wish to learn more about our strategic recruitments, you can read interviews with them here.
Old riddles
The first day focused on medicine and solving old questions. The second session was themed “Lost and found in life science” and was started by keynote Walther Parson, Medical University of Innsbruck. His group works within two tracks. The first is to identify remains of individuals who have died in war, natural disasters or other circumstances where identification might be difficult. The other track is DNA identification of historical persons, which involves the same tools as in crime investigations.
One of the examples he mentioned was the famous German poet Schiller. The identification of Schiller has been an interesting case, not least because he had two skeletons assigned to him for almost 100 years. By extensive DNA analyses of relatives to Schiller it was finally concluded that none of the skeletons belonged to him.
The sessions of the first day also covered the evolutionary loss and gain of animal societies, individualized cancer systems medicine, and Clinical impact of recurrent coding and non-coding mutations in Chronic lymphocytic Leukemia.
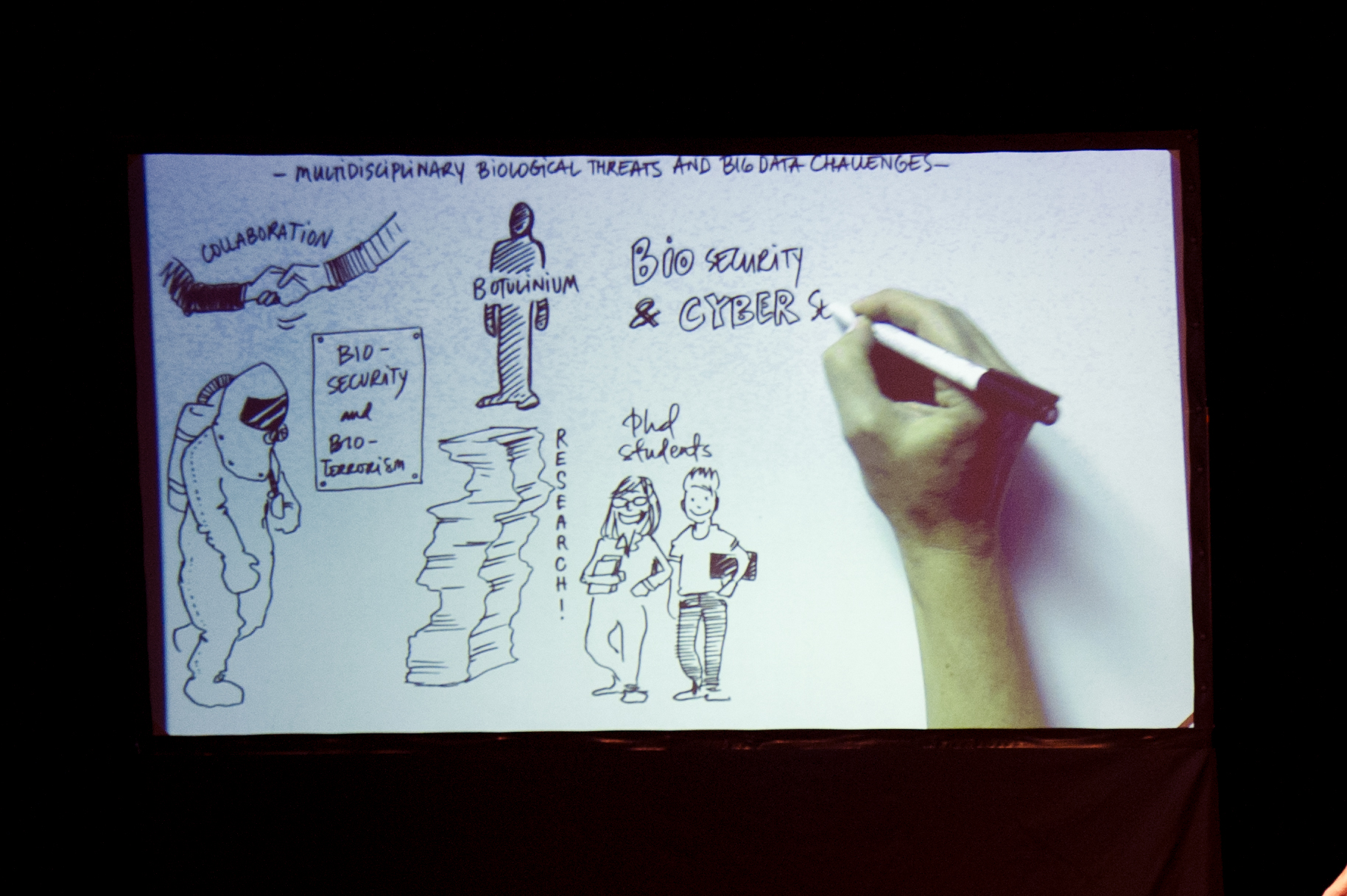
All of the speakers were accompanied by live drawings that illustrated their talk.
Water and soil
The second day was dedicated to environmental research. The aim was to exchange information and ideas on how environmental sciences can benefit from the tools, infrastructure and expertise in molecular biosciences offered by SciLifeLab. The day started out with a number of case studies from ecosystem-centered research. Both terrestrial and aquatic ecosystems were covered, with emphasis on the tools used.
In the afternoon the participants were divided into smaller groups to discuss which methods they would like to see SciLifeLab offer in the future. The ideas were presented in a plenary discussion at the end of the day. Some of the suggestions and requests were:
- More resources for training and courses, including mentoring by the platforms.
- Broaden the amplicon sequencing service by including steps before the actual sequencing, e.g. PCR of amplicons.
- Even more standardized methods, where applicable.
Full lists of speakers and participants for both days are available at the event page.
Photos
Live drawing, illustrating Rickard Knutsson’s talk on Multidisciplinary biological threats and big data challenges
Mingle
Carolina Wählby at Fellows corner
Mingle
Walther Parson, giving his talk on DNA identification of famous people