New accurate method for pathway enrichment analysis
Researchers from SciLifeLab have developed a new network-based Pathway enrichment analysis method called ANUBIX. The new method is considerably more accurate than previous network crosstalk based methods and was recently described in Nature Scientific Reports.
During recent years, big improvements in molecular biology have led to a large increase in high-throughput data of, among other things, lists of differentially expressed genes or proteins. These lists can be used to identify genes with important roles during certain conditions.
More insight about the biological mechanisms is often needed though, such as information about how functional gene sets are related to genes in the result list. The study of this relation, between target genes and functional gene sets (pathways) is called pathway enrichment analysis.
By using Pathway enrichment analysis it is possible to find out which biological processes are affected by altered gene activities under specific conditions. So far, it has been a challenge to develop a method that can efficiently avoid false positives while keeping a high sensitivity. Previous methods generally assume that a pathway (a group of partly linked genes) have the same statistical properties as a random gene set.
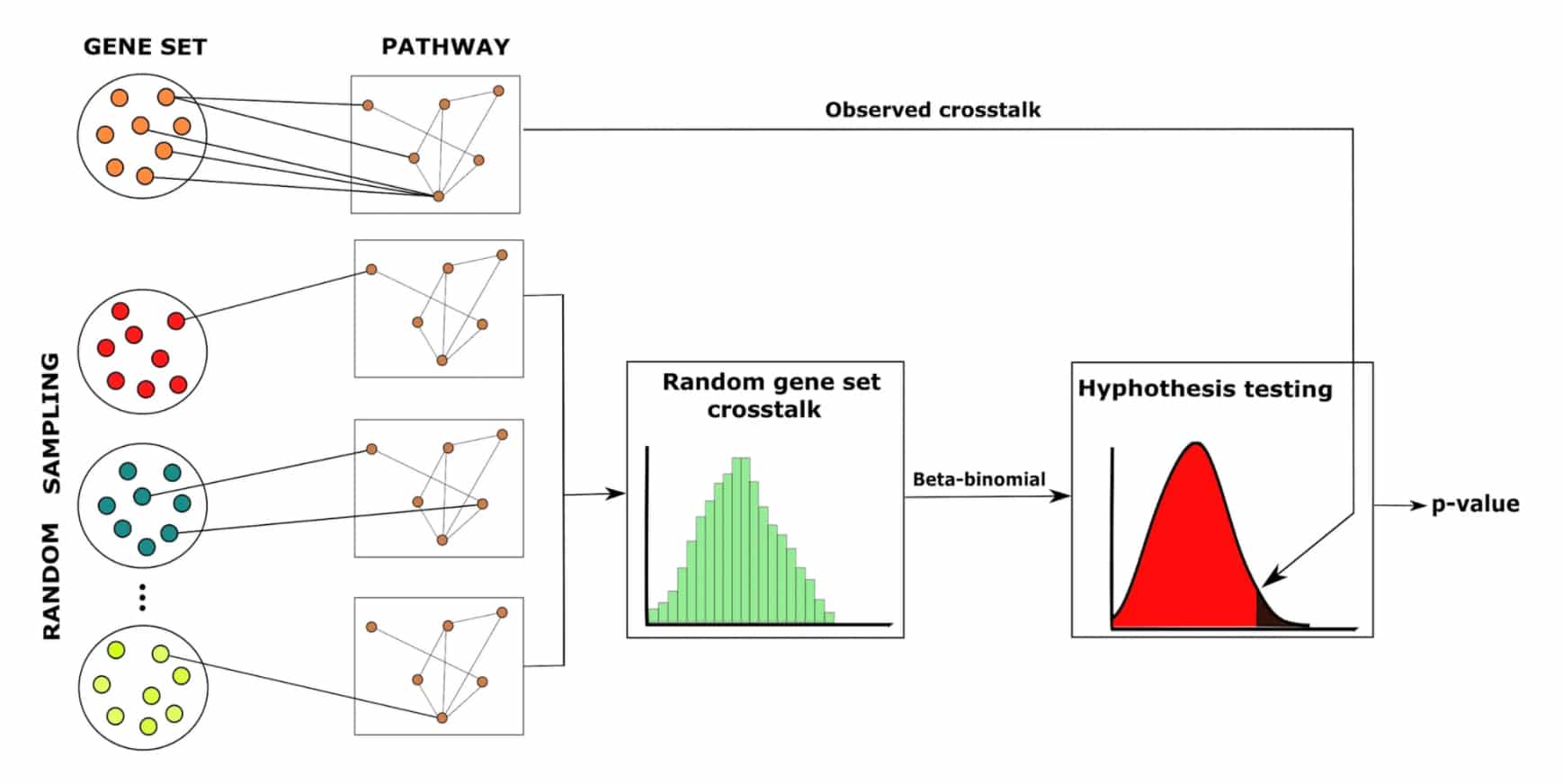
In this study, led by SciLifeLab researcher Erik Sonnhammer (Stockholm University), researchers developed a new network-based method, called ANUBIX, based on sampling random gene sets against intact pathways. The new method takes into account that genes in a pathway often have more internal connections than randomly picked genes.
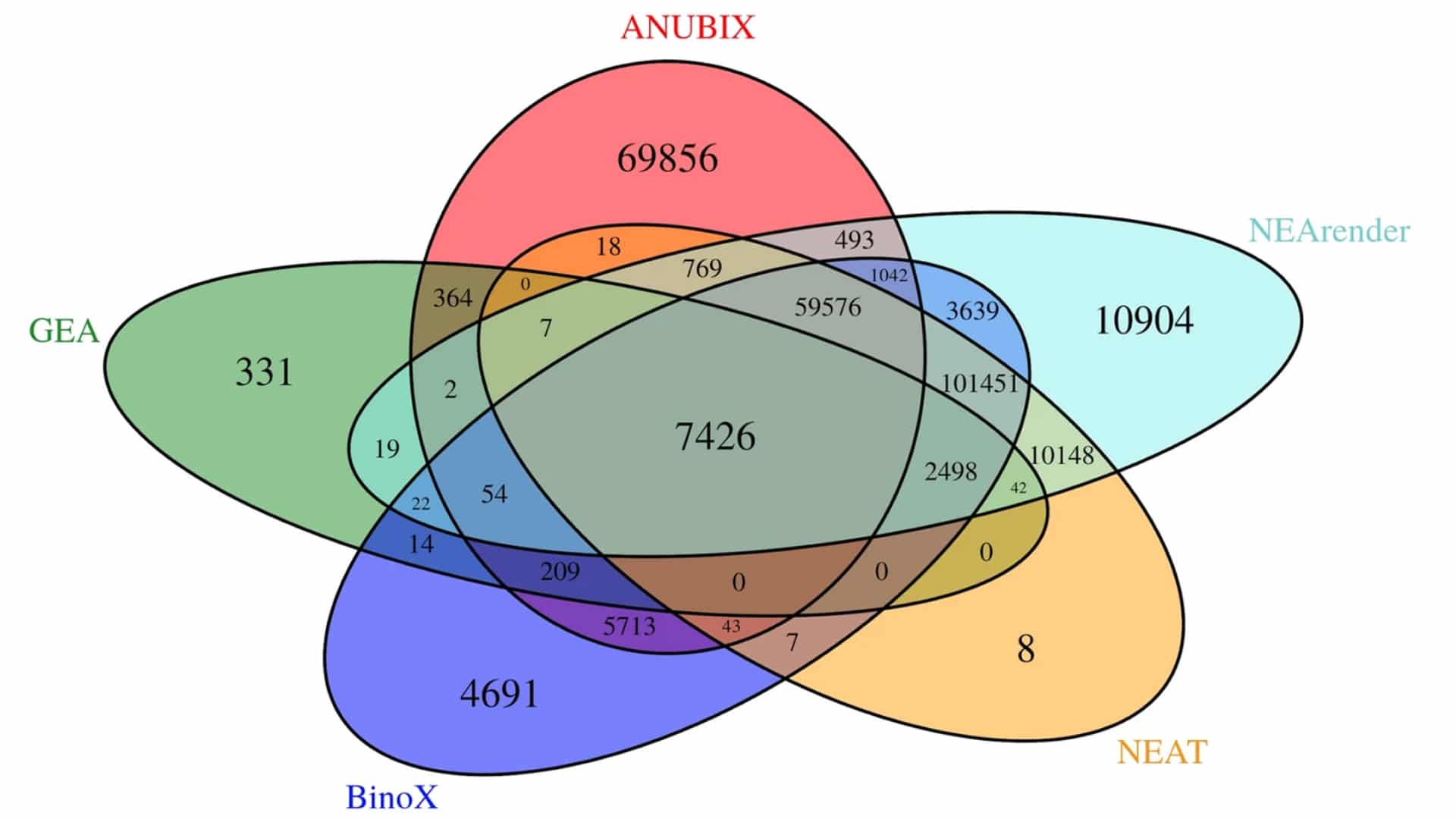
Benchmarking shows that ANUBIX is considerably more accurate than previous network crosstalk based methods, which have the drawback of modelling pathways as random gene sets. The researchers were able to demonstrate that ANUBIX does not have a bias for finding certain pathways, which existing methods do, and provide evidence that ANUBIX can find biologically relevant pathways that are missed by other methods.