New bioinformatics pipeline reveals non-canonical proteins
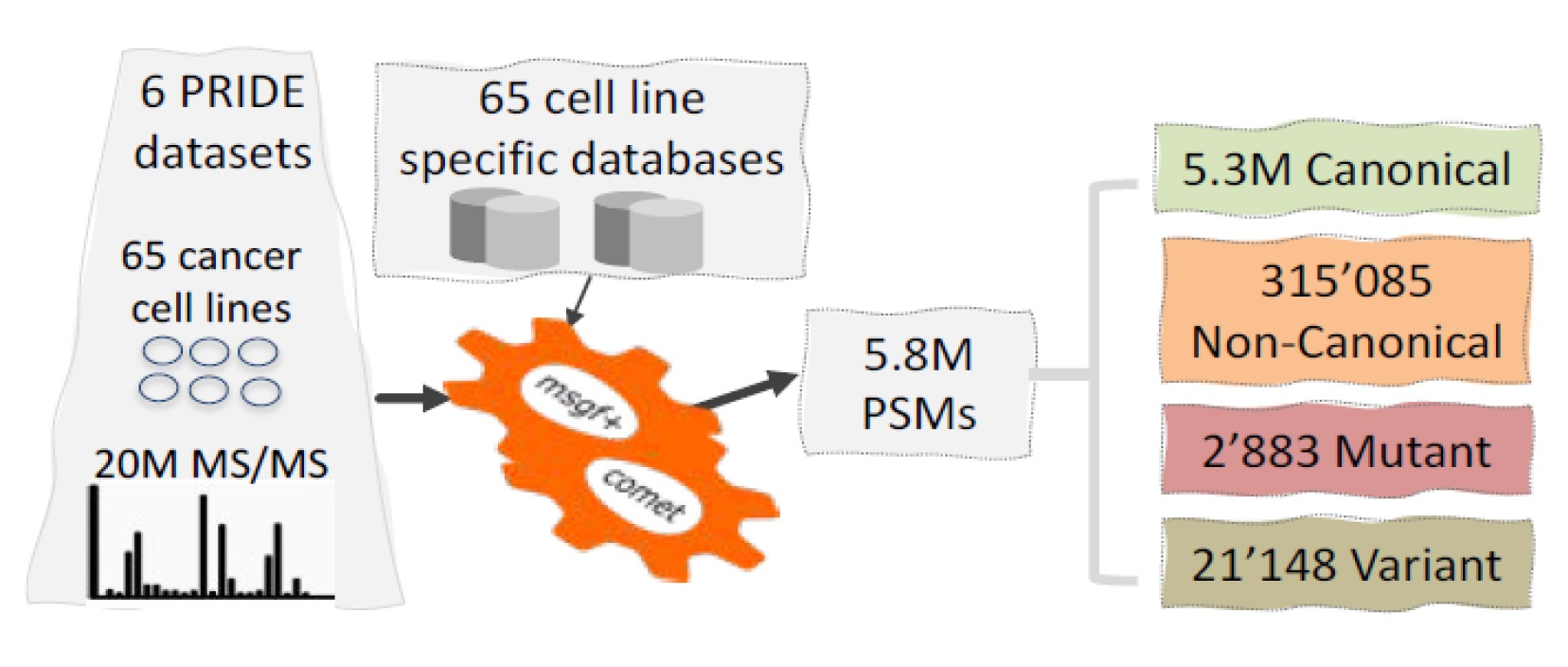
Researchers from SciLifeLab and EMBL-EBI, Cambridge, have developed a new bioinformatics pipeline, enabling users to identify non-canonical proteins – unusual protein products from non-coding genomic regions that are often found in tumors – from local or public proteomic datasets.
In recent years, the field of Proteomics, and its subdomain Proteogenomics has discovered a group of cryptic, almost mysterious proteins, dubbed non-canonical proteins. Non-canonical proteins are unusual protein products not part of the annotated protein-coding genome repertoire. They arise from unexpected translation of otherwise non-coding genomic regions, or from mRNA splicing aberrations, or from out-of-frame translation.
Whereas certainly some of these non-canonical proteins are expected to have genuine functions in the context of a healthy cell or tissue, it is becoming clear that more of these cryptic proteins are popping up in abnormal contexts such as cancer. Indeed, recent publications are indicating that some of the non-canonical proteins are specific to cancer cells.
It is therefore of paramount importance to be able to detect non-canonical proteins so as to investigate any potential tumoral/pathogenic function these proteins may have and consider them as tumor-specific targets potentially useful in the context of targeted Immunotherapy.
In a collaborative effort between SciLifeLab proteogenomics unit and Janne Lehtiö’s lab (SciLifeLab/Karolinska Institutet) and the group of Yasset Perez-Riverol at EMBL-EBI, Cambridge, highlighting the SciLifeLab-EMBL memorandum of understanding, researchers have implemented a Bioinformatics pipeline that enables any user to mine a local or public proteomics dataset and discover these non-canonical protein products. The study is now published in Bioinformatics.