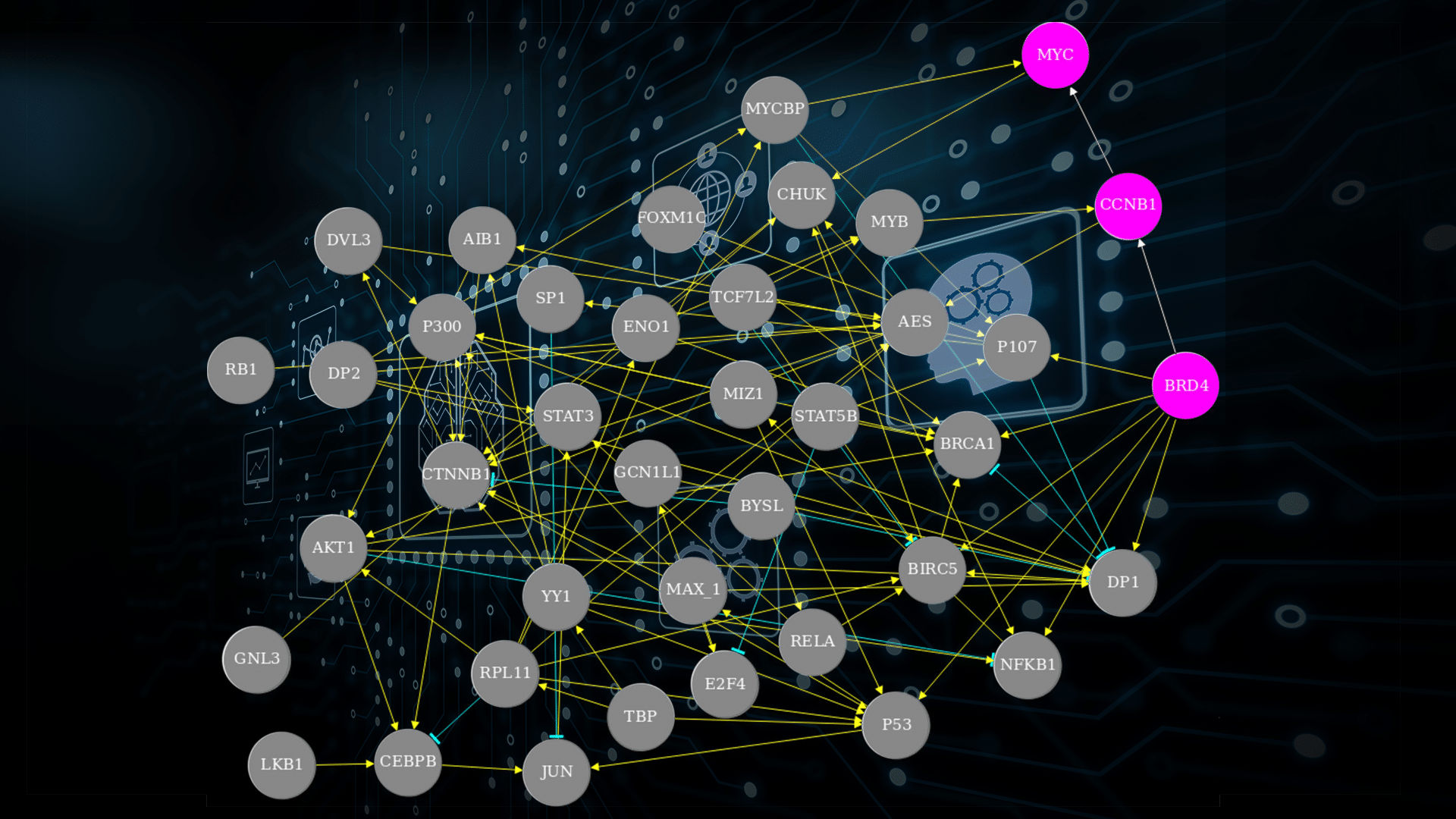
New gene regulatory network unravelling oncogenic mechanisms validated by novel approach
In a recent study, led by Erik Sonnhammer (SciLifeLab/Stockholm University), researchers constructed a new, highly reliable Gene Regulatory Network (GRN), relevant for cancer research. Furthermore, they created a system that can be used to determine the accuracy of theoretical models describing GRN:s.
Cell proliferation and cell death are controlled by specific regulatory systems, and cancer can be seen as an altered state of such a system. Researchers have over time proposed many models of different GRN:s, and the success of such models can be measured by how well they are able to predict the response of the system during specific scenarios.
So far, amongst all the inference techniques used to create GRN:s, using knock-down of genes and measuring the system’s response has proved to show the greatest accuracy. However, relatively few such experiments have been conducted by scientists, compared to the vast landscape of possible regulatory interactions that could occur in a cell. Because of this, researchers have had a hard time inferring correct GRN:s that mirrors reality.
To unravel oncogenic mechanisms this study first collected a set of genes, mostly regulators associated with cancer and thought to interact to some degree with the oncogene MYC. These genes were individually knocked down, after which the response in the cells’ gene expression was measured. Using this data, a GRN was constructed. However, without a gold standard to compare against it is difficult to assess how accurate a GRN is.
Because of this, the team has created a new method for inferring whether a GRN is reliable or not, and their technique can be applied to GRN:s inferred with any method.
The motivation behind the study was that nobody has derived a regulatory network for these genes as a system before. The hardest part for the team lied in performing the experiments as well as in designing the method to assess predictiveness.
“The main finding of our study was a gene regulatory network relevant to cancer, which was derived by carrying out knockdown perturbation experiments in a cancer cell line. The network was shown to be highly reliable by a novel approach to assess predictiveness against validation data. Furthermore, it unravels a number of previously unknown oncogenic mechanisms”, says Eriks Sonnhammer (SciLifeLab/SU).
In the study, published in Nature Scientific Reports, the researchers tested their assessment method on several alternative GRN:s and concluded that: “Almost all inferred GRN:s were more predictive than expected by chance, and some were vastly more predictive. These top-performing GRN:s were also able to predict an independent pairwise-gene perturbation validation dataset significantly better than expected by chance. The best GRN contained many known links as well as proposes many novel links, two of which were verified experimentally”.
“Our findings propose new key genes in cancer regulation and could lead to new therapies. It could shift the research focus within the MYC oncogene field, a large subfield of cancer research, as well as for other oncogenes”, says Erik Sonnhammer.