New microscopy approaches reveal the transcription factor sliding mechanism
Researchers from SciLifeLab and Uppsala University have developed new microscopy approaches that make it possible to visualize the mechanism with which DNA-binding proteins search the genome for their specific binding sites.
Stored inside the nucleus, our DNA relies on the binding of different transcription factor proteins to regulate genes. Failure to bind to the correct transcription factor binding site could be disastrous for the cell, leading to expression of genes that should be turned off or no expression at all when translation is vital.
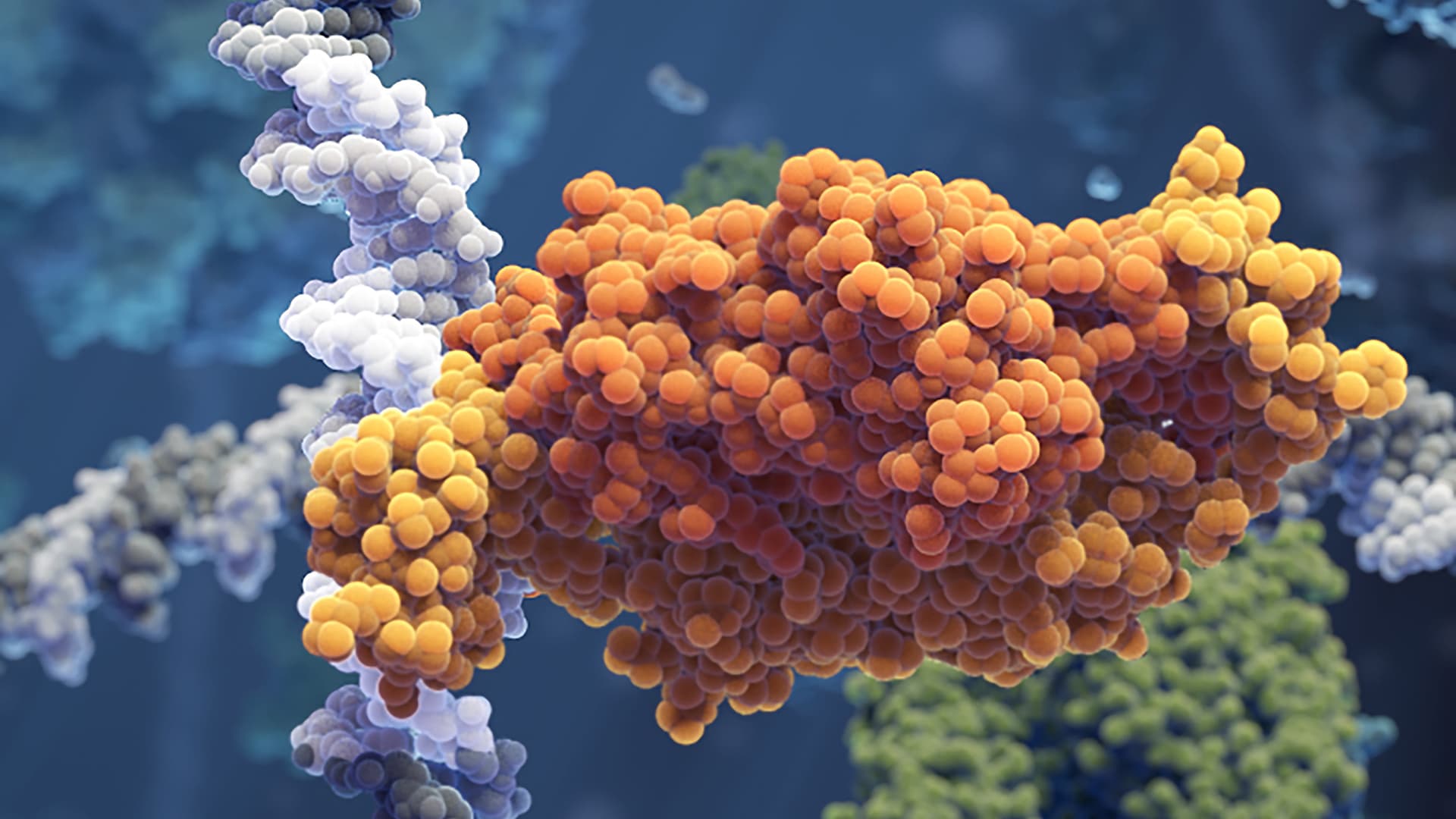
The specific binding sites are tiny compared to the entire genome and how the transcription factors manage to find them has been studied extensively. Many transcription factors seem to utilize a process called facilitated diffusion in which they alternately diffuse back-and-forth across DNA segments in one dimension and diffuse in 3D space to find new segments to search in case they fail to locate the right sequence. The 1D sliding mechanism has so far remained poorly understood.
In a new study, led by Sebastian Deindl and Johan Elf (SciLifeLab/Uppsala University), researchers were able to shed new light on the sliding mechanism.
“The molecular mechanism of the scanning process has been poorly understood, and it has remained a great mystery how transcription factors manage to slide fast on non-specific DNA sequences, yet at the same time bind efficiently to specific targets”, says Emil Marklund (Uppsala University), PhD student and joint first author, in a press release from Uppsala University.
The two research teams developed new fluorescence microscopy imaging approaches that made it possible to observe individual transcription factors sliding along the DNA molecule in real time as they search for and bind to the correct site.
“It is exciting that we were able to develop new imaging approaches to directly observe, for the first time, if and how often the sliding protein fails to recognize and slides past its binding site,” says Sebastian Deindl.
Due to the high scanning speed of the sliding transcription factor, which scans 10 base pairs every 100 microseconds, the researchers had to develop a new way of shooting and tracking microscopy movies at an extremely high speed in order to visualize the movement.
“It’s great that we can push the dynamic observation of bimolecular interactions to the sub-millisecond time scale – this is where the chemistry of life happens,” says Johan Elf.
The results, published in Nature, show that the sliding transcription factors are quite sloppy and regularly miss their target sites. The researchers could also observe that the sliding transcription factors frequently made short hops instead of strictly following the helical geometry of the DNA molecule.
“By hopping, the protein trades thorough scanning for speed, so it can scan DNA faster. This is a really smart choice by the protein, since it will find the target twice as fast using this search mechanism,” says Emil Marklund.
Photo: Visual Science, Johan Elf, Irmeli Barkefors