New technique enables fine mapping of closely related cells in situ
SciLifeLab researchers have developed a new technique called “Probabilistic cell typing by in situ sequencing” or pciSeq. The new technique can identify cell types as well as determine their spatial location in a much more efficient way than previous methods.
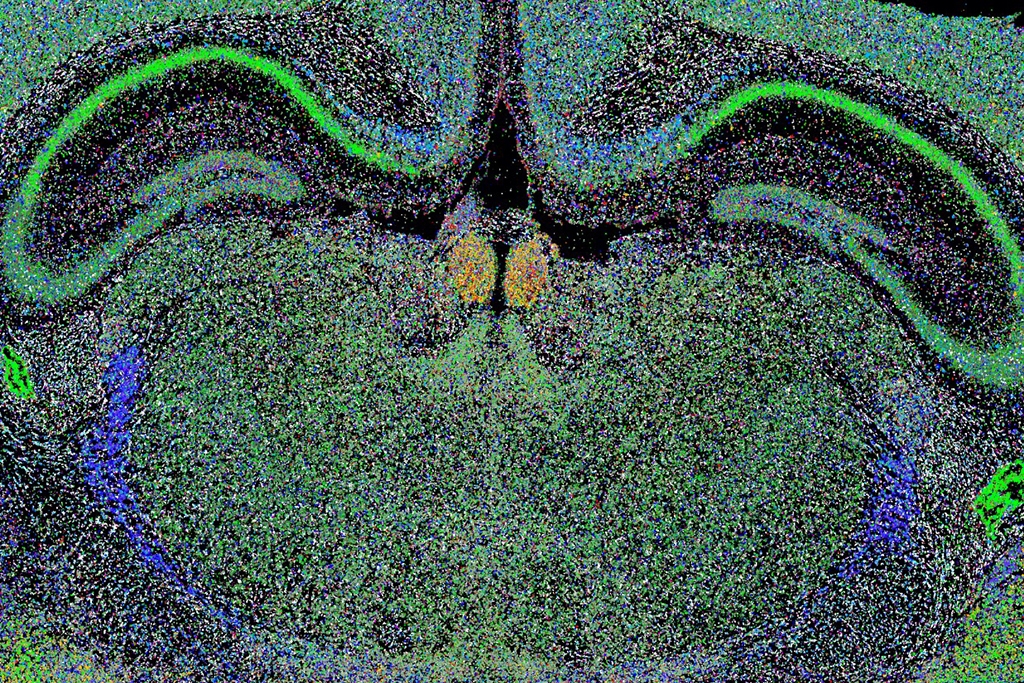
The global project “Human cell Atlas” has reached a point where it is now possible to catalog virtually all human cell types based on their molecular content rather than how they look in a microscope. In order to understand how different tissues function, we also need to create maps of the spatial organization of the different tissue cell types, however.
In a recent study, led by Mats Nilsson (SciLifeLab/SU) researchers presented an effective way to deal with the problem demonstrating the strength of the technique by mapping neurons in the mouse hippocampus CA1 and cortex. The map is the most complete map of all nerve cells in the mouse brain so far. The study was published in Nature Methods.
The main reason such great resources are spent on cataloging and mapping human organs in the “Human cell Atlas-project” is that if you know the molecular content of our different cell types, it will be much easier to understand the differences between healthy and diseased tissue at the cellular and molecular level. Armed with that knowledge, it will most likely be easier to find effective therapies against complex diseases such as psychiatric, neurodegenerative, and cancer. PciSeq could help speed up this process.