SciLifeLab researchers collaborate to introduce Expansion Spatial Transcriptomics
A team of researchers from SciLifeLab and Stanford University recently published an exciting new paper in Nature Methods detailing their work on Expansion Spatial Transcriptomics (Ex-ST). Array-based spatial transcriptomics techniques are widely used to analyze gene expression in tissues. However, their ability to provide detailed information is limited by the density of the arrays. Ex-ST overcomes this limitation by first clearing and expanding the tissue, followed by capturing the entire polyadenylated transcriptome using an improved protocol, thereby surpassing the previous resolution limitations.
In recent years, many approaches for spatial gene expression profiling have been developed. The pioneering technique targeting the entire transcriptome was introduced by Ståhl et al. in 2016 and is now commercialized by 10x Genomics as the Visium platform.
“In this work, we combined tissue expansion and spatial transcriptomics to increase the resolution of the 10x Genomics Visium assay,” says SciLifeLab researcher Žaneta Andrusivová.
Ex-ST uses two distinct poly-deoxythymidine probes to capture transcripts. One probe is attached to the gel matrix, while the other is a spatially barcoded probe positioned on the surface of the Visium capture array. This sophisticated approach has been designed to provide a clearer picture of gene expression within tissues and tissue structures.
“This exciting work demonstrates the possibility of re-engineering and expanding tissue to overcome one of the challenges with current commercial solutions of spatial transcriptomics, enabling the capture of gene activity in single cells,” says Molecular Biotechnology professor Joakim Lundeberg.
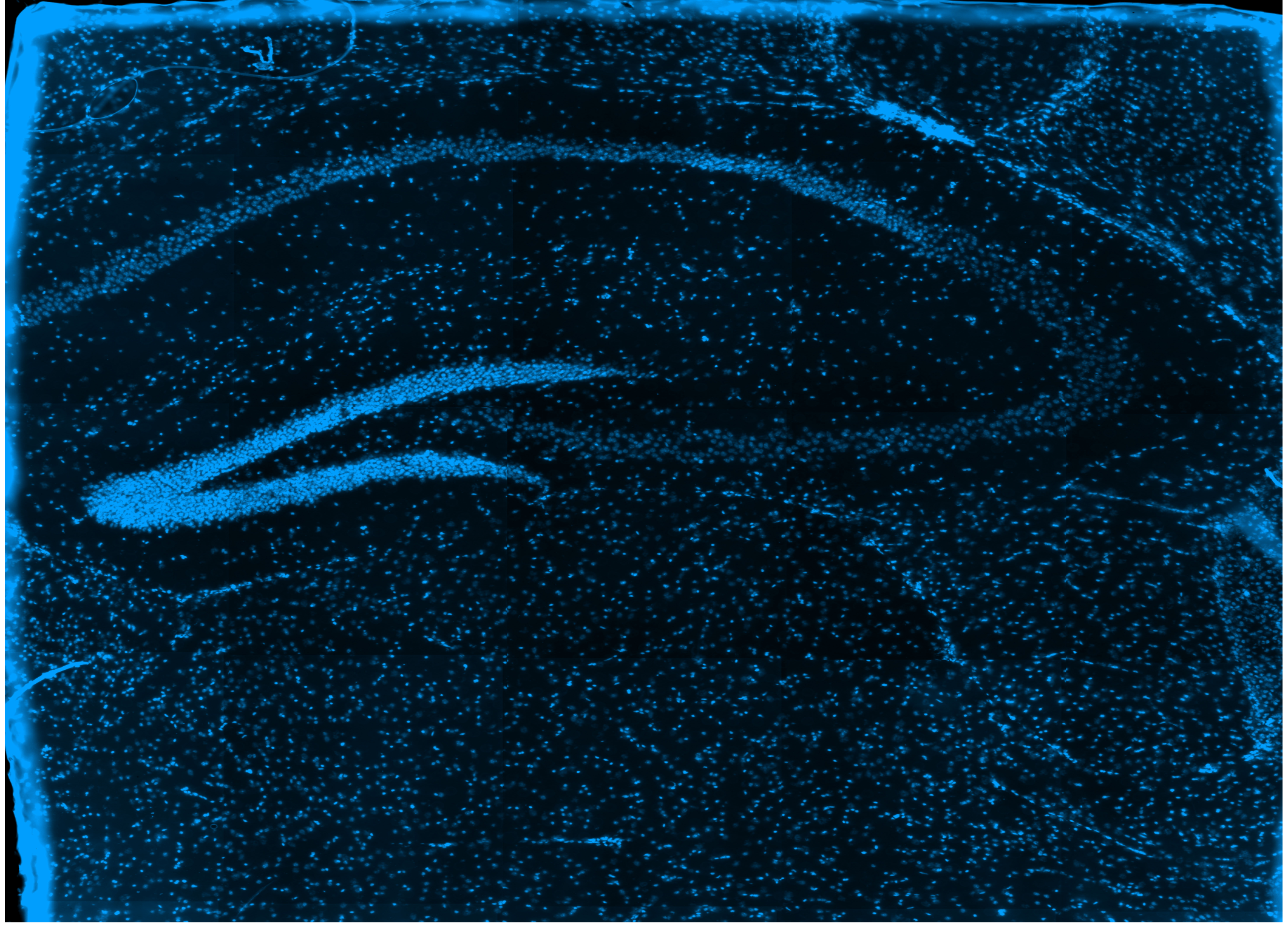
To demonstrate their method, the researcher used the mouse brain’s olfactory bulb and hippocampus regions chosen for their distinct anatomical and molecular features. Results indicated that while Ex-ST might capture fewer unique molecules and genes per spatial feature, its ability to study gene expression within tissue structures in greater detail is commendable.
In conclusion, the introduction of Ex-ST in the field of spatial transcriptomics suggests a promising path forward. While it’s still in its early days, the method’s potential to provide deeper insights into tissue organization is evident.