SciLifeLab researchers win protein structure competition
A research group at SciLifeLab, led by Jens Carlsson at Stockholm University, has won a worldwide competition to predict the three-dimensional structure of a G protein-coupled receptor using computer modeling. Their results are published in two articles in the scientific journal Structure.
G protein-coupled receptors (GPCRs) have key functions in many physiological processes and close to one third of all pharmaceuticals act through these receptors.
The aim of the GPCR DOCK competition is to evaluate the accuracy of 3D structural models of GPCRs and their interactions with drug molecules. Researchers from all over the world are challenged to predict how small molecules, such as pharmaceuticals, bind to GPCRs at the atomic level. The experimentally determined 3D structure of the GPCRs are kept secret until the participating research groups have submitted their models.
The GPCR DOCK 2013 competition involved prediction of the structures of serotonin receptors type 1B and 2B bound to an anti-migraine drug. Out of hundreds of submitted models, Jens Carlsson’s group contributed with the best prediction for the serotonin 1B receptor and was also among the highest ranked for the 2B-receptor. The first article in Structure summarizes the results of the GPCR DOCK competition.
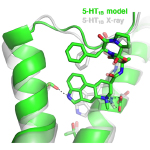
The model and experimentally determined structures are shown in green and white, respectively.
Accurate modeling of how small molecules bind to GPCRs could contribute to development of new pharmaceuticals. Many receptors have very similar binding pockets and it is important to develop drugs that bind selectively to avoid side effects. In the second article, Jens Carlsson’s group demonstrates that selective drugs can be designed based on the experimentally determined serotonin receptor structures, but that further improvement is required to accomplish this based on models.
“Our results show that we can predict the overall binding mode of a drug to a GPCR, but that it is still very challenging to capture subtle differences between closely related receptors. Still, as 3D structures for more than 90% of all GPCRs remain unknown, models of this accuracy could have a significant impact on drug development.”, says Jens Carlsson.
Publications:
Advances in GPCR modeling evaluated by the GPCR Dock 2013 assessment: meeting new challenges. Kufareva, I.; Katritch, V.; GPCR Dock 2013 Participants; Stevens, R. C.; Abagyan, R. Structure (2014) In press.
Structure-based Discovery of Selective Serotonin 5-HT1B Receptor Ligands. Rodríguez, D.; Brea, J.; Loza, M. I.; Carlsson, J. Structure (2014) 22, 1–12.




