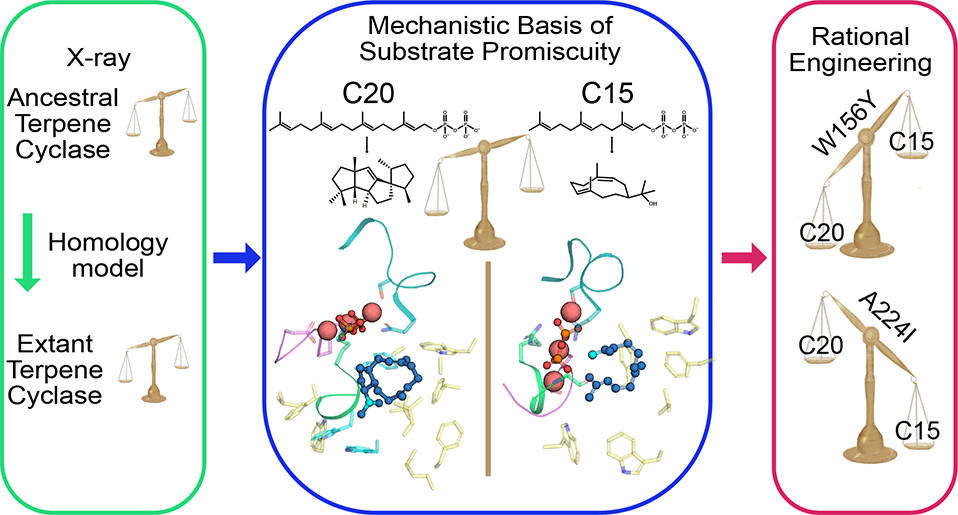
Using ancestral proteins to deal with common X-ray crystallography limitations
Structural information is vital when it comes to understanding biochemical processes and in fields, such as enzyme engineering, the information is used to reveal the catalytic mechanisms in detail. In order to gather this information, researchers are using X-ray crystallography, but the technique has its limitations and often works poorly with proteins that are unstable or have a limited solubility.
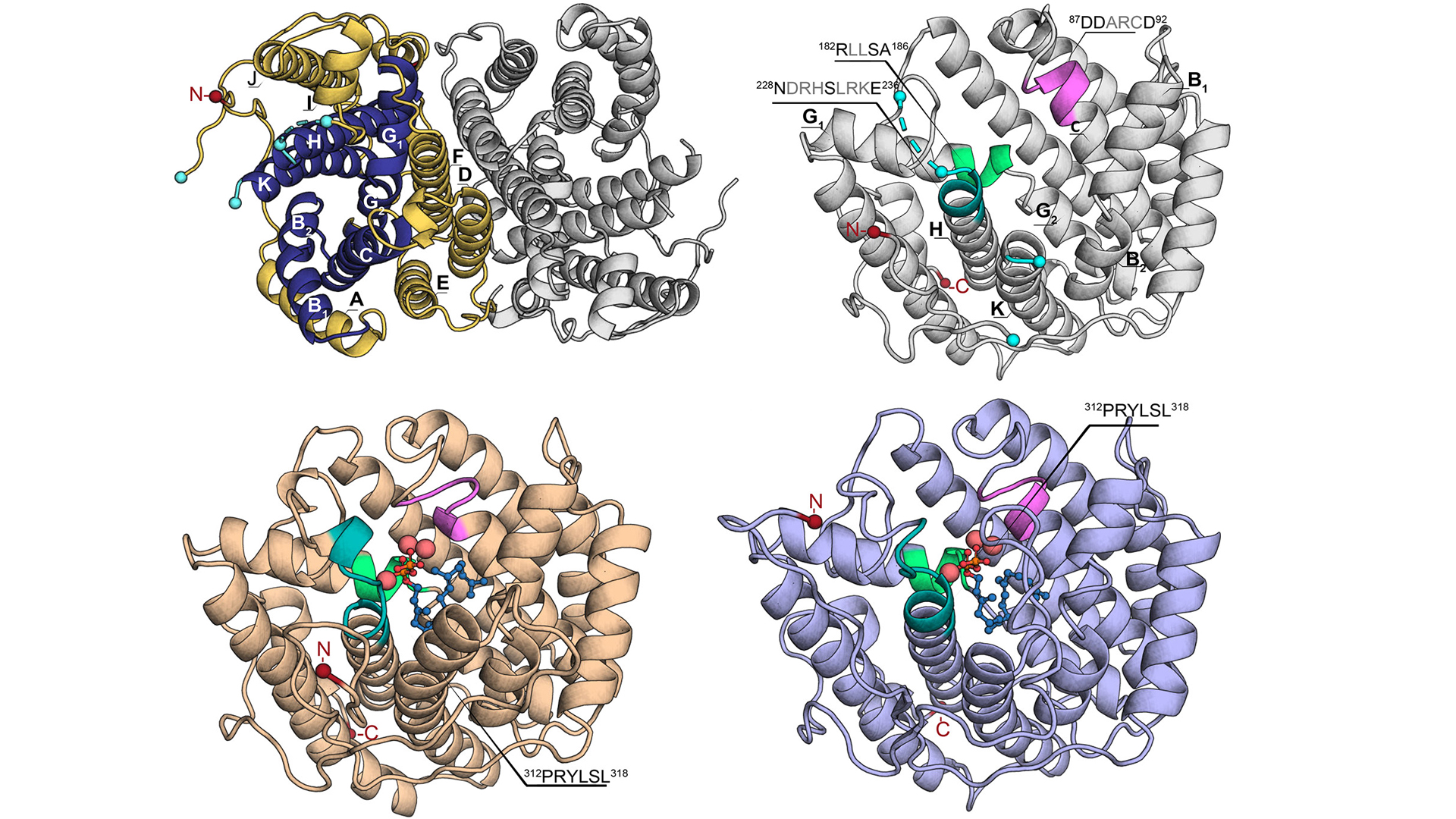
To overcome this major obstacle, SciLifeLab researcher Per-Olof Syrén KTH) and his team, including first author Karen Schriever (KTH), and researchers from Karolinska Institutet, have developed a method that is based on the design and engineering of stable ancestral proteins with high homology and substrate specificity to existing enzymes. Information of modern proteins and enzymes that can resist crystallization was also utilized in the process.
The results and potential of the method was published in the journal of J. Am. Chem. Soc. (JACS) and the research was also featured on the cover. The method could have a broad application scope in chemical biology and provide access to structures of biomolecules that are otherwise challenging to study as well as in generating highly active and stable scaffolds for biophysical characterization and biotechnological applications.
“This work shows the potential of learning from the past to understand and engineer the present”, says Per-Olof Syrén.