Research interests
One branch of our research regards development of algorithms and methods in bioinformatics that can be used to answer biomedically interesting problems. Another branch regards studies of gene/protein families, including structural aspects, in order to arrive at knowledge about their function and at the same time contributing to classification of parts of the huge protein world.
One of our ongoing projects regards investigation of differences at the genetic and proteomic levels between the two main types of heart failure (HF) — HF with reduced left ventricular ejection fraction (HFrEF; “the large dilated heart”) and HF with preserved left ventricular ejection fraction (HFpEF; “the small stiff heart”). This is part of the large study PREFERS, where genomics, proteomics, metabolomics and advanced imaging tools are used in an integrative fashion in order to clarify pathophysiologic mechanisms of HFpEF and HFrEF. The project is a collaboration with Cecilia Linde and Hans Persson at Karolinska Institutet and is funded by Astra-Zeneca.
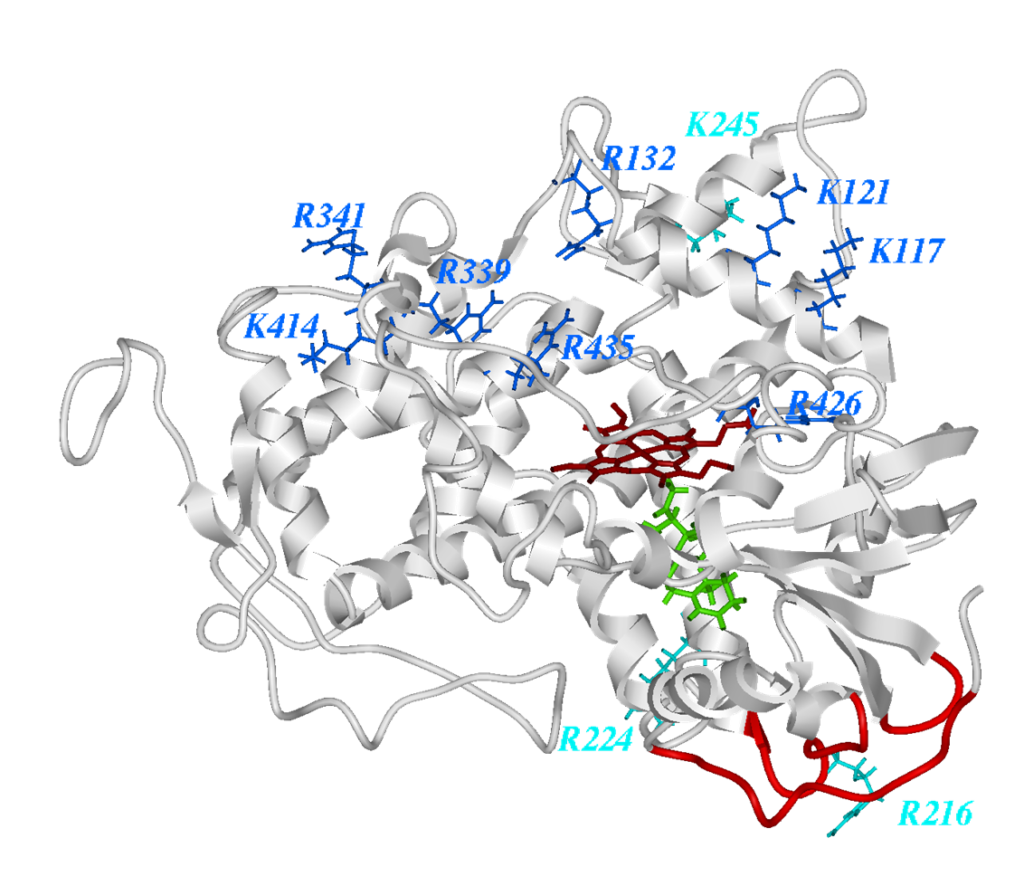
Another project focuses on developing and using structural calculations to investigate mutational effects and for functional characterisation. With structural calculations techniques also considering protein stability in addition to measurements around the active site and analysis of conserved residues, the effect of a mutation can in many cases be predicted with good confidence.
Furthermore, we are working on protein classification, where we use hidden Markov models (HMMs) for functional assignments of protein families. We develop an automated strategy for such sub-classification tasks, making it possible to utilise these methods on large scale analyses. We have an emphasis on the two very large families SDR and MDR (short-chain and medium-chain dehydrogenases/reductases, respectively), which have been shown to be of central metabolic importance for all organisms.
Bengt Persson is also director of the SciLifeLab Bioinformatics Platform and Bioinformatics Short-term Support and Infrastructure (BILS).
Group members
Bengt Persson, prof., bengt.persson@scilifelab.se
Sarbashis Das, post-doc, sarbashis.das@icm.uu.se
Christoffer Frisk, project assistant, christoffer.frisk@icm.uu.se
Linus Östberg, Ph.D. student, linus.ostberg@scilifelab.se