We aim to discover and understand the molecular mechanisms underpinning how health status is affected by exposures, such as food, microbiota and environmental pollutants. We work on data from both population-based cohorts as well as human dietary intervention studies, with a strong focus on data-driven life science, including nutritional and molecular epidemiology. To understand regulation of the metabolic system in relation to exposures and health, we work with different Omics data, mostly reflecting metabolites, proteins and microbiota.
To perform molecular- and mechanism-oriented research, we actively contribute to the development of freely available data analytical tools. We typically combine our MUVR algorithm for machine learning analysis with linear models to identify Omics variables-of-interest that can be either biomarker candidates for exposure or health assessment.
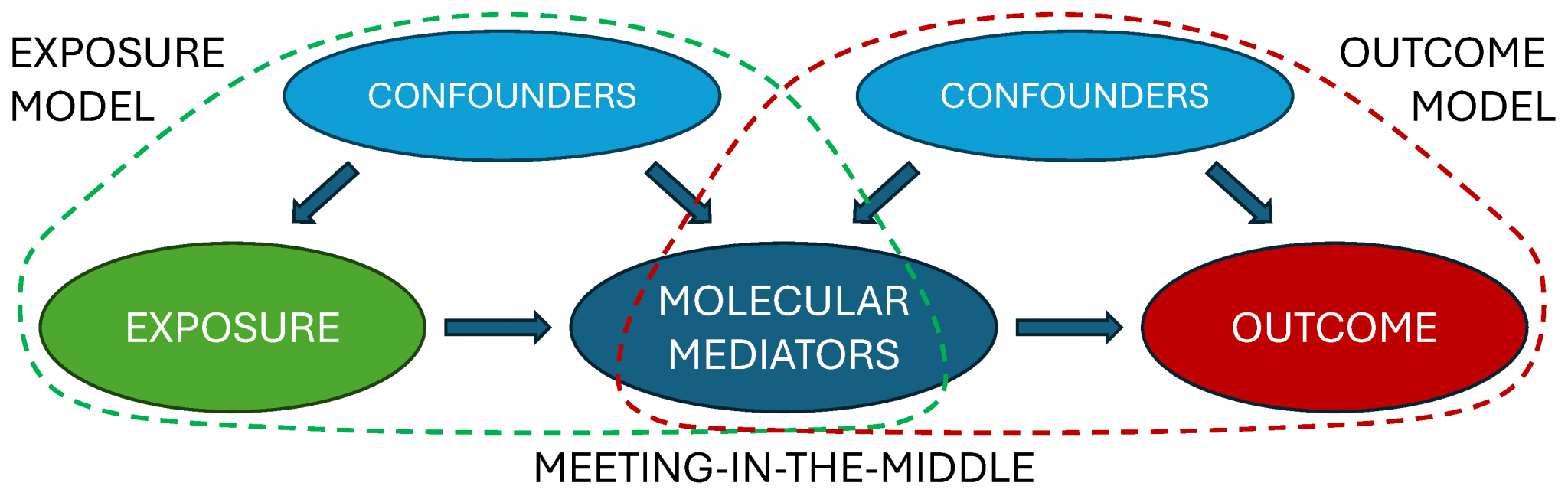
We have also become fond of the “meet-in-the-middle” approach for molecular epidemiology, where potential mediators can be identified from the intersection of Omics variables related to both exposures and outcomes. We can then visualize and interpret how these potential molecular mediators link e.g. diet or pollutant exposures to type 2 diabetes, cardiovascular disease or other health conditions using our triPlot tool.
In addition, Dr Brunius is Scientific advisor to the Chalmers Mass Spectrometry Infrastructure which performs mass spectrometry-based metabolomics analysis as part of the SciLifeLab Metabolomics platform. He supplies the infrastructures with computational tools to monitor and improve data quality as well as to improve automation and reproducibility in the pre-processing of instrument data into actionable format for down-stream data analysis. This involvement highlights the effective interaction between data generation and data usage for life science research.
Group Members:
Carl Brunius, Associate Professor
Yingxiao Yan, doctoral student
Elise Nordin, doctoral student
Olle Hartvigsson, doctoral student
Anton Ribbenstedt, postdoc