Research Interests
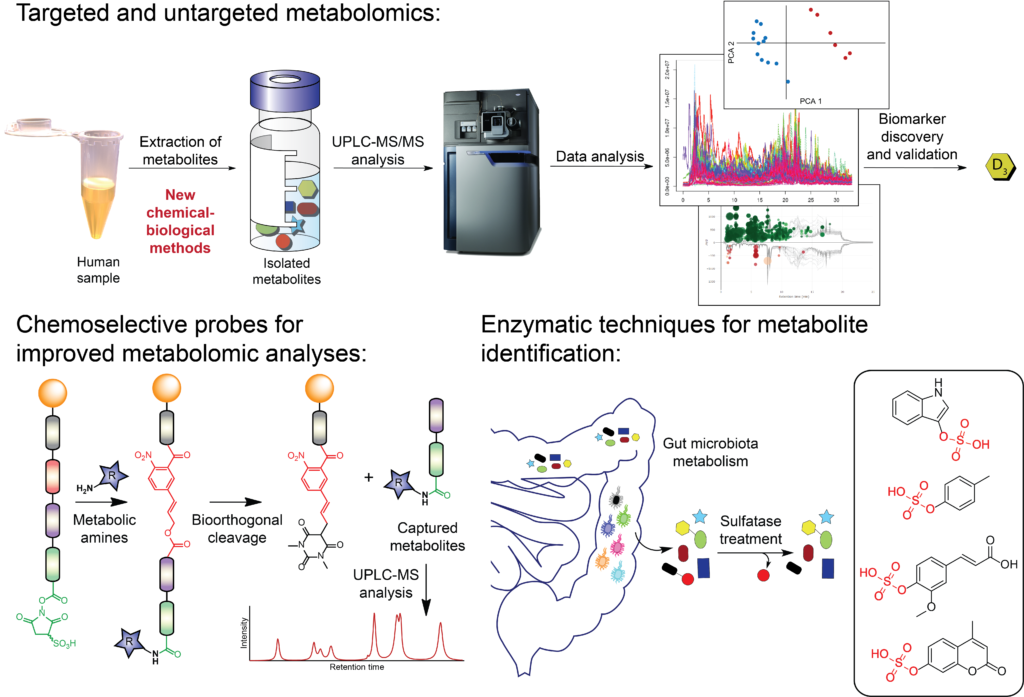
The Globisch lab is an international research group with a focus on the development of new Chemical Biology-based methodologies to improve the analysis of small molecule metabolites in biological samples. These new methodologies are aimed at enhancing the scope of metabolomics-based research. In our multidisciplinary research we work at the interface of Chemistry and Biology with a combination of Organic Chemistry, Chemical Biology techniques, Biochemistry, mass spectrometric analysis of metabolites, and metabolomics. The discovery of specific early-stage biomarkers, new drug targets and the development of new therapeutic interventions are crucial for disease prevention and management towards personalized medicine.
Biomarker Discovery is a challenging and multidisciplinary task. We are developing new tools for mass spectrometric analysis and are focussed on elucidating microbiota metabolism in the human host towards the discovery of unknown bioactive metabolites. We investigate any human sample type, e.g. fecal, urine, and plasma samples from pancreatic cancer patients, colorectal cancer, gut-brain axis, and other common diseases.
Group members
- Daniel Globisch, Associate Professor
- Weifeng Lin, PhD student
- Ioanna Tsiara, PhD student
- Sydney Mwasambu, PhD student
- Vladyslav Dovhalyuk, PhD student
- Carina de Souza Anselmo, postdoctoral researcher
- Fan Yang, postdoctoral researcher
- Pauline Seeburger, Internship student
- Marko Spasic, Master’s student
- Hien Kim Thi Le, Master’s student
- Anaïs Fournier, Erasmus+ exchange student
- Vincent Ripoll, Erasmus+ exchange student
- Athanasia Byzyka, Internship student