Whole genome sequencing from just six nuclei
Enabled by the SciLifeLab Microbial Single Cell Genomics unit (MSCG), National Genomics Infrastructure (NGI) and National Bioinformatics Infrastructure (NBIS), researchers from Uppsala University have developed a new and more effective way to sequence genomes from organisms that are otherwise hard to sequence.
The reasons why some organisms are harder to sequence are usually because of their small size, their inability to grow in a lab environment or because they grow in conjunction with other sometimes similar species making it hard to distinguish between their DNA.
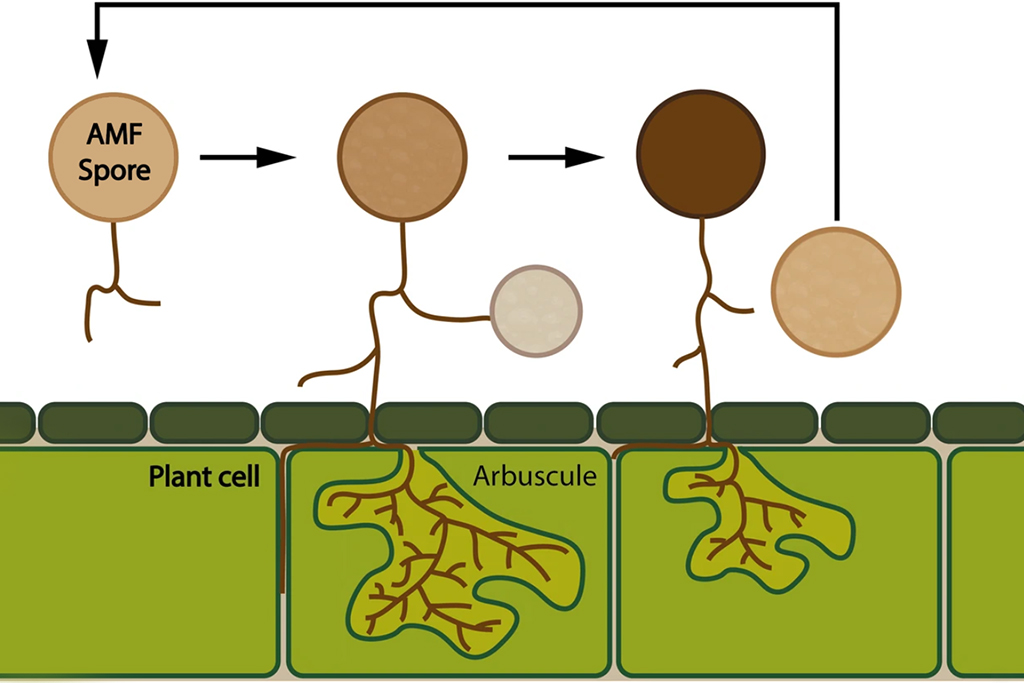
In a recent study, researchers from Uppsala University, including last author Anna Rosling and first author Merce Montoliu-Nerin, was able to sequence an arbuscular mycorrhizal fungi, a group of plant symbionts that complete their life cycle below ground, by using nuclear sorting from a single spore.
These fungi are important plant symbionts that improve plant health and productivity of plants such as crops or fruit trees. Arbuscular mycorrhizal fungi have a cryptic life cycle and produce multinucleate asexual spores, which makes it a suitable model for this kind of method development.
The result, published in Nature Scientific Reports, showed that a comprehensive genome assembly could be produced from as little as six to seven nuclei. According to the researchers, the method is highly applicable for a broad range of organisms and will greatly improve the ability to study other multicellular eukaryotes with complex life cycles.
Read the press release from Uppsala University.