Method developed to infer accurate GRNs from uninformative gene expression data
A group of researchers from Taiwan, Sweden and the USA led by SciLifeLab researcher Erik Sonnhammer (SciLifeLab/SU), presents a new general method for accurately inferring Gene Regulatory Networks (GRNs) from sets of uninformative gene expression data.
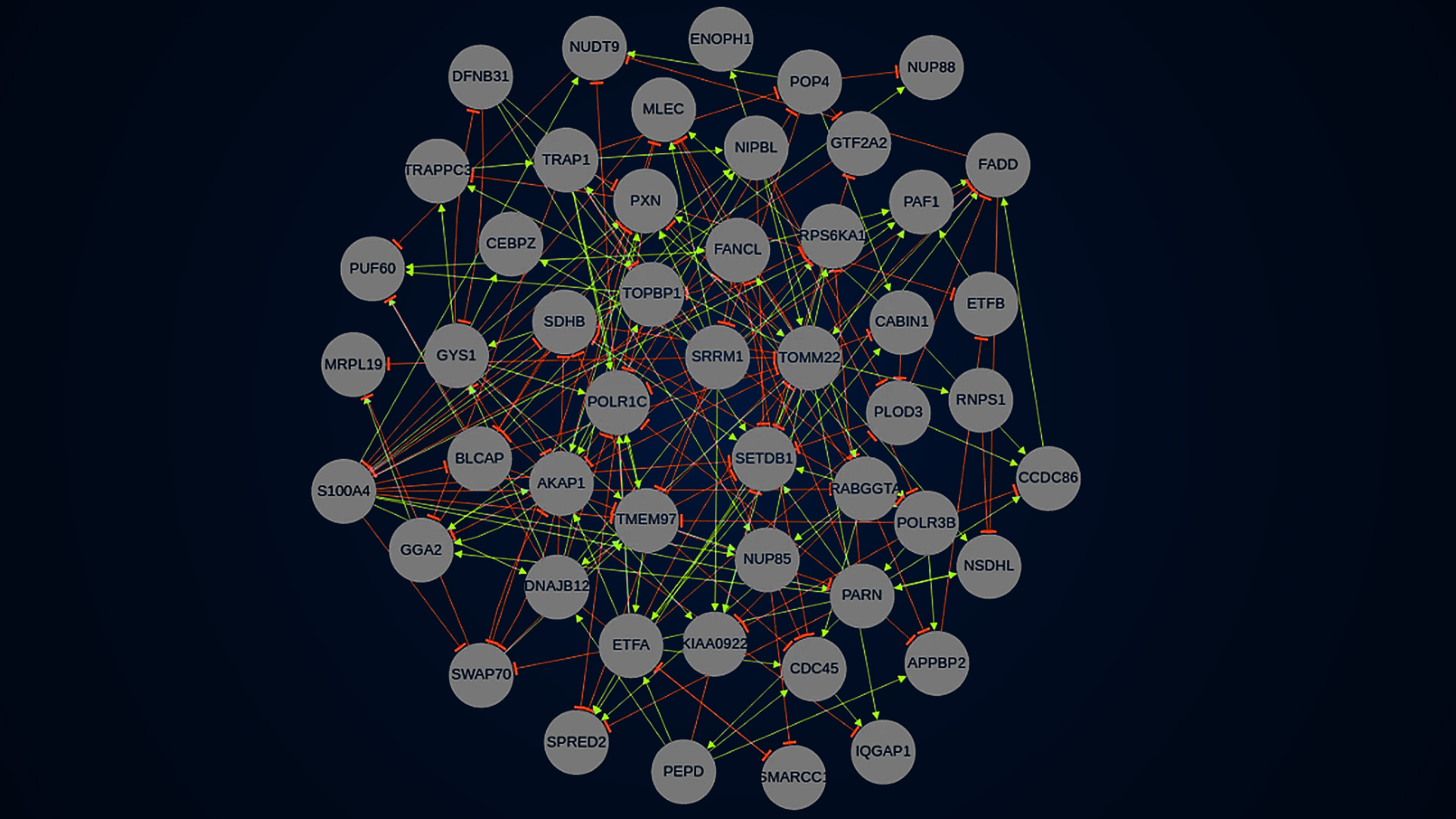
In the article, published in Nature, the researchers describe a novel pipeline for inferring Gene Regulatory Networks. By perturbing genes it is possible to gather data on their gene expressions. This perturbation-based gene expression data, based on interactions among components of living cells, can then be used to infer so-called Gene Regulatory Networks (GRNs).
“Such networks are useful for providing mechanistic insights on a biological system.”, says last author Erik Sonnhammer.
The researchers present a new method that can be applied to gene expression datasets to infer accurate GRNs. Erik Sonnhammer (SciLifeLab/SU) explains the main findings of their research.
“We describe a new gene reduction pipeline that eliminates uninformative genes from gene expression data, as well as its application to public cancer cell line data”.
The method can be applied by researchers to their own datasets to infer accurate GRNs.
“Most gene expression datasets have a very low signal-to-noise ratio (SNR) level, causing them to be too uninformative to infer accurate GRNs. With our method, other scientists can eliminate uninformative genes from the data and infer accurate GRNs from the remainder.”, says Erik Sonnhammer.
Which part of this research was the hardest?
“Figuring out how to detect the most uninformative genes”, he concludes.