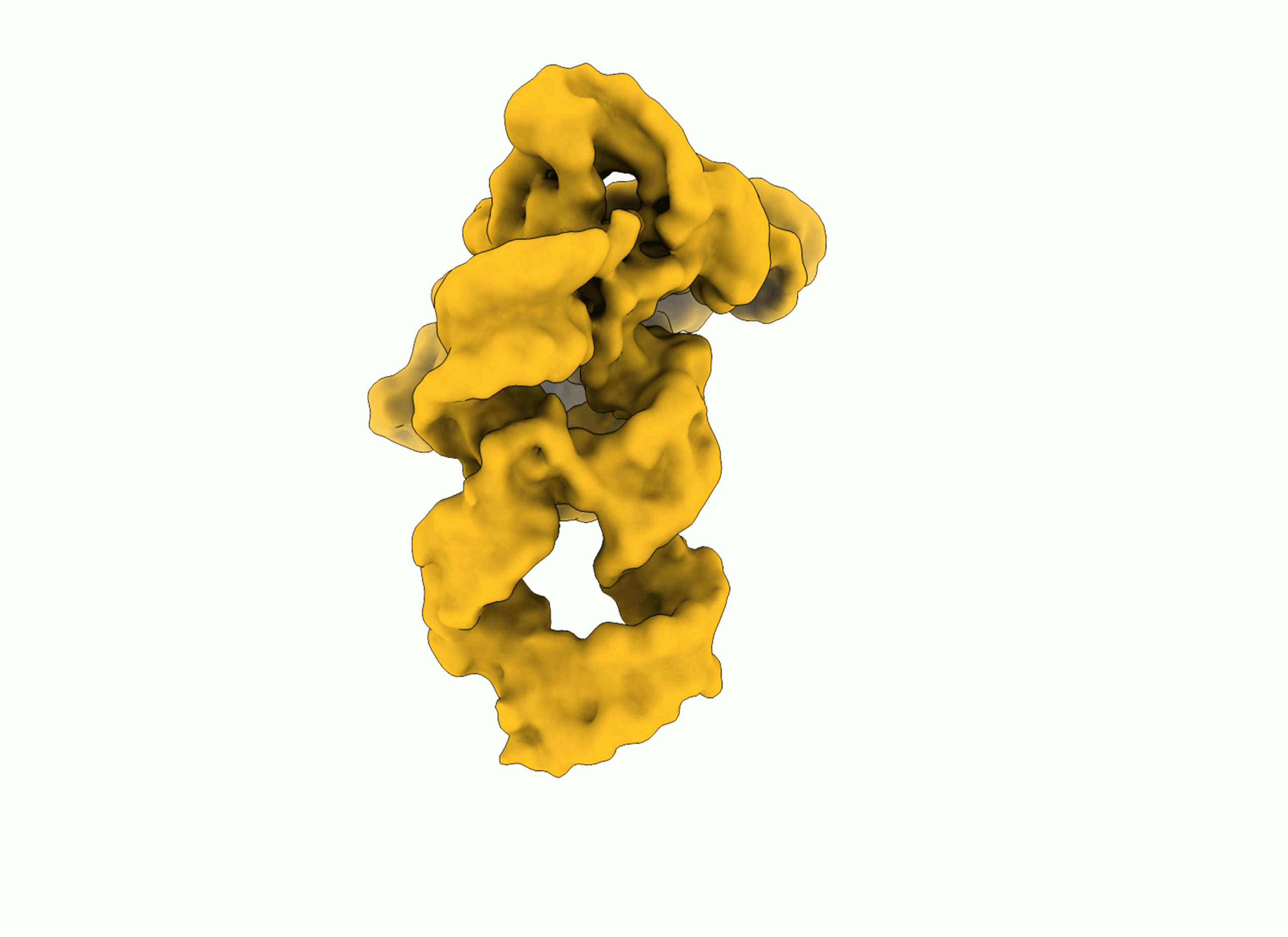RNA in action: revealing a ribozyme’s self-assembly
Researchers have visualized, in unprecedented detail, how a large RNA molecule assembles itself into a functional machine, revealing a dynamic choreography that helps better understand structural folding and misfolding, almost frame by frame.
How RNAs avoid folding errors and evolve into complex cellular machines is now revealed by researchers in an international collaboration, including organizations such as SciLifeLab, EMBL Grenoble, Center for Structural Systems Biology (CSSB) Hamburg and the Istituto Italiano di Tecnologia (IIT). The study is published in Nature Communications.
Watch the video of the RNA at our LinkedIn page.
Marco Marcia’s group at SciLifeLab and Uppsala University shows how a large RNA molecule folds and assembles itself.
“Structural biology typically captures static photographs of biological molecules. Here, we pushed the limits of cryo-EM and image processing to capture a dynamic movie of our target,” says Marco Marcia and continues, “Achieving this result implied a lot of optimization of our sample, of the technique, and even the development of new data analysis pipelines. An important factor for success was to establish exciting new scientific collaborations along the way”.
This work paves the way for AI-driven RNA prediction, a step towards an ‘AlphaFold for RNA’.
Notoriously difficult target
RNA is a central biological molecule, now widely harnessed in medicine and nanotechnology. Like proteins, RNA often gets its function from its three-dimensional structure. Researchers have recorded how a large RNA molecule folds, flexes, and assembles itself into a functional biological machine, almost frame by frame.
The study used an ensemble of state-of-the-art techniques such as cryo-electron microscopy (cryo-EM), small-angle X-ray scattering (SAXS), RNA biochemistry and enzymology, image processing, and molecular simulations. By doing so, scientists have, for the first time, captured the dynamic process by which a self-splicing ribozyme folds into its functional structure.
“Determining RNA structures is a challenging task. The inherent flexibility and negative charge make RNA a notorious target for structural studies. Persistent efforts and extensive screening on electron microscopes ultimately led us to visualise elusive RNA dynamics,” says Shekhar Jadhav, SciLifeLab and Uppsala University postdoc, and first author of the study.
The result is the most complete description to date of an RNA building itself, revealing how it avoids the biological equivalent of outtakes: misfolded, non-functional states known as kinetic traps.
Insights into early RNA-based life
Group II introns, the ribozymes analyzed in the study, are thought to be the ancestors of the spliceosome, the complex machinery that edits RNA in human cells. By revealing how these molecules fold efficiently and avoid kinetic traps, the study provides new insight into how early RNA-based life may have evolved its editing tools.
This work sets the stage for RNA design and engineering, which can guide how future biotechnologies might script RNA molecules to fold correctly for use in therapeutics or nanobiotechnology.
Opening the Door to RNA AI
“This work is expected to play a key role in shaping artificial intelligence approaches to RNA structure prediction, paving the way towards a new ‘AlphaFold for RNA,” says Marco Marcia.
This convergence of experimental precision and machine learning marks a new phase for RNA structural biology, where AI and cryo-EM can learn from each other to predict, visualize, and understand the dynamics of life’s most versatile molecule.
Watch the video at our LinkedIn page.
Learn more in the EMBL article
DOI: 10.1038/s41467-025-65502-8
Top image: Shekhar Jadhav/EMBL, frame from a GIF grabbed by SciLifeLab.