Structural basis of mitochondrial translation
Researchers from SciLifeLab, led by Fellow Alexey Amunts (Stockholm University), have been able to provide functional details about the mitochondrial translation by analyzing mitoribosomes with bound messenger and transfer RNA, and nascent polypeptide. The two studies, published in eLife and Nature Communications, reveal new mitoribosome components and suggest how the expression of our genetic information is coordinated in mitochondria.
Gene expression in humans takes place in the cytosol and mitochondria. In mitochondria, it involves transcription and translation of genes responsible for the maintenance of the cellular energy balance. The mitoribosome is a key player in this process, and its impaired activity results in 1 out of ~4000 individuals experiencing myopathies and neurodegenerative diseases.
In two new studies, from SciLifeLab Fellow Alexey Amunts’ (SU) lab, researchers have utilized a structural approach in an effort to explain the molecular mechanisms behind the process.
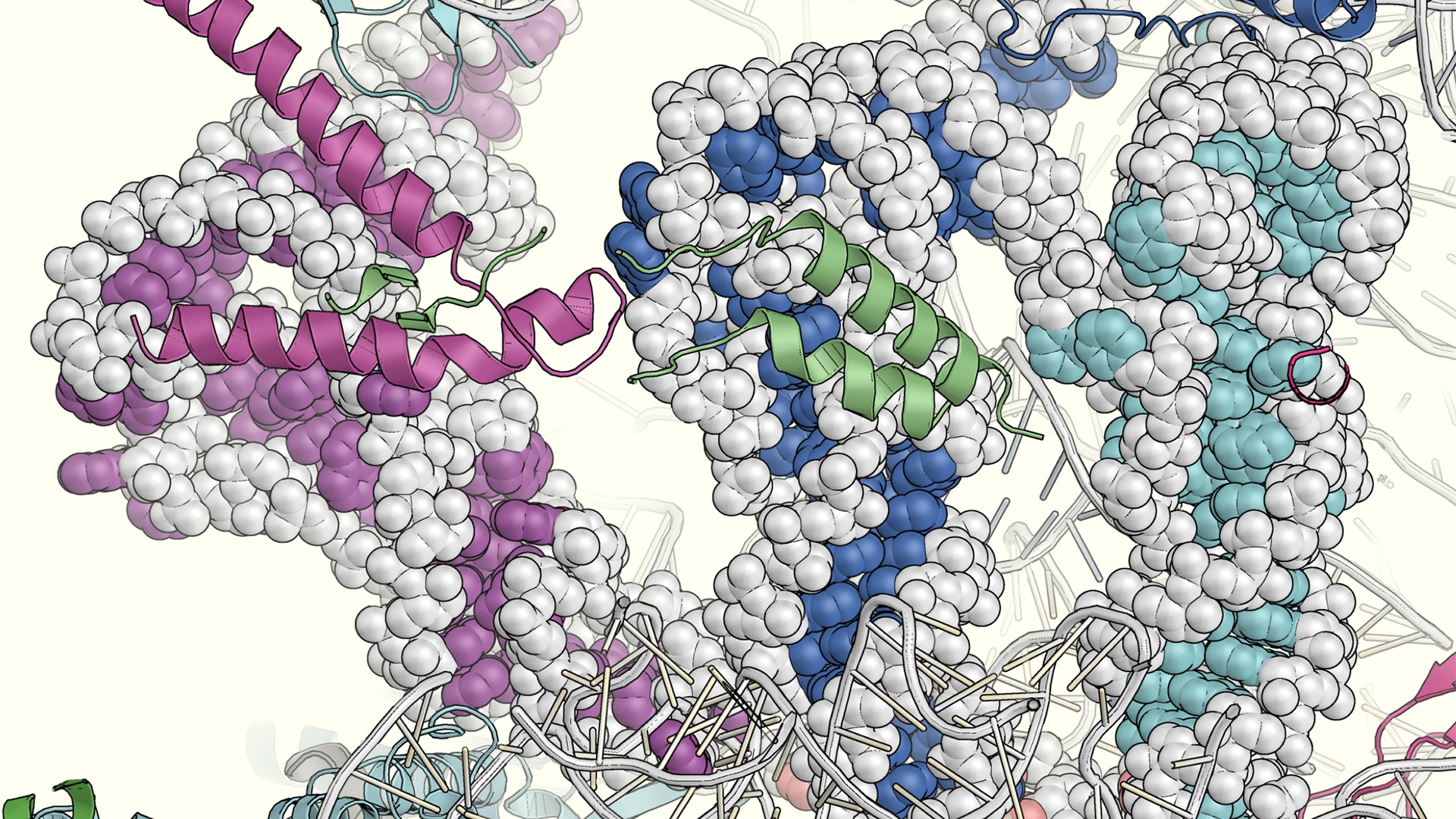
The first study illuminates how gene expression is coordinated in human mitochondria. The analysis revealed eight structures with compositional and conformational variations, which could be linked to previously unknown specific protein components guiding transfer RNA movements, as the messenger RNA is being translated.
When mitoribosomes are activated, an additional module is found, associated with a dedicated platform leading to the messenger RNA channel. It was identified as the LRPPRC-SLIRP complex that delivers transcribed genetic information into the mitochondrial matrix, facilitating translation by handing the messenger RNA over to the mitoribosome.
This newly discovered link between transcription and translation in mitochondria provides a potential explanation to why mutations in LRPPRC can cause Leigh syndrome, a severe neurological disorder.
Due to the limited resolution of the complex isolated from human cells, the second study was designed to obtain a higher resolution of the system by using a fungal model organism, Neurospora crassa, instead. Cryo-EM data from the study allowed the researchers to build the most complete model of the mitoribosome available so far. It features mitochondrial codon-anticodon base pairings between messenger and transfer RNA, for the first time, and provides a trace of the entire ligand path through the mitoribosome.
Future research will focus on how the complex multi-component mitochondrial translation machinery is assembled.