National Facility for Exposomics
The National Facility for Exposomics at SciLifeLab is coordinated by Stockholm University and the Department of Environmental Science (ACES). With expertise in small molecule mass spectrometry in human and environmental samples [1-4], we operate as Infrastructure Unit of the SciLifeLab Metabolomics Platform.
The exposome was first described in 2005 to encompass all environmental exposures throughout the lifetime, and exposomic techniques continue to evolve with a purpose to better characterize what we are exposed to and how these exposures interact as risk factors for disease. [5-7] The current infrastructure is dedicated to study the chemical fraction of the exposome which is increasingly accessible by high resolution mass spectrometry. Our methodological approaches and research directions were recently highlighted in a multidisciplinary perspective. [8]
Applications
Our mission is to make state-of-the-art methods in high resolution chemical exposomics available to a wide user base of national and international researchers who wish to understand exposure and effects of complex organic chemical mixtures through analysis of human, wildlife, or environmental samples. We continually adapt best available technologies and workflows to meet demands of our users for targeted and untargeted chemical exposomics in a range of sample types, including air and water, and a range of biofluids and tissues [1-4]. Our general working philosophy is that the data processing workflows should be achievable with open science tools to fully support data FAIRness, as demanded by the national research audience today.
References
[6] Miller G.W., Exposome: a new field, a new journal, Exposome, 1 (1), 2021
[8] Zhang P. et al., Defining the Scope of Exposome Studies and Research Needs from a Multidisciplinary Perspective. Environ. Sci. Technol. Letters, 8 (10), 2021
Services
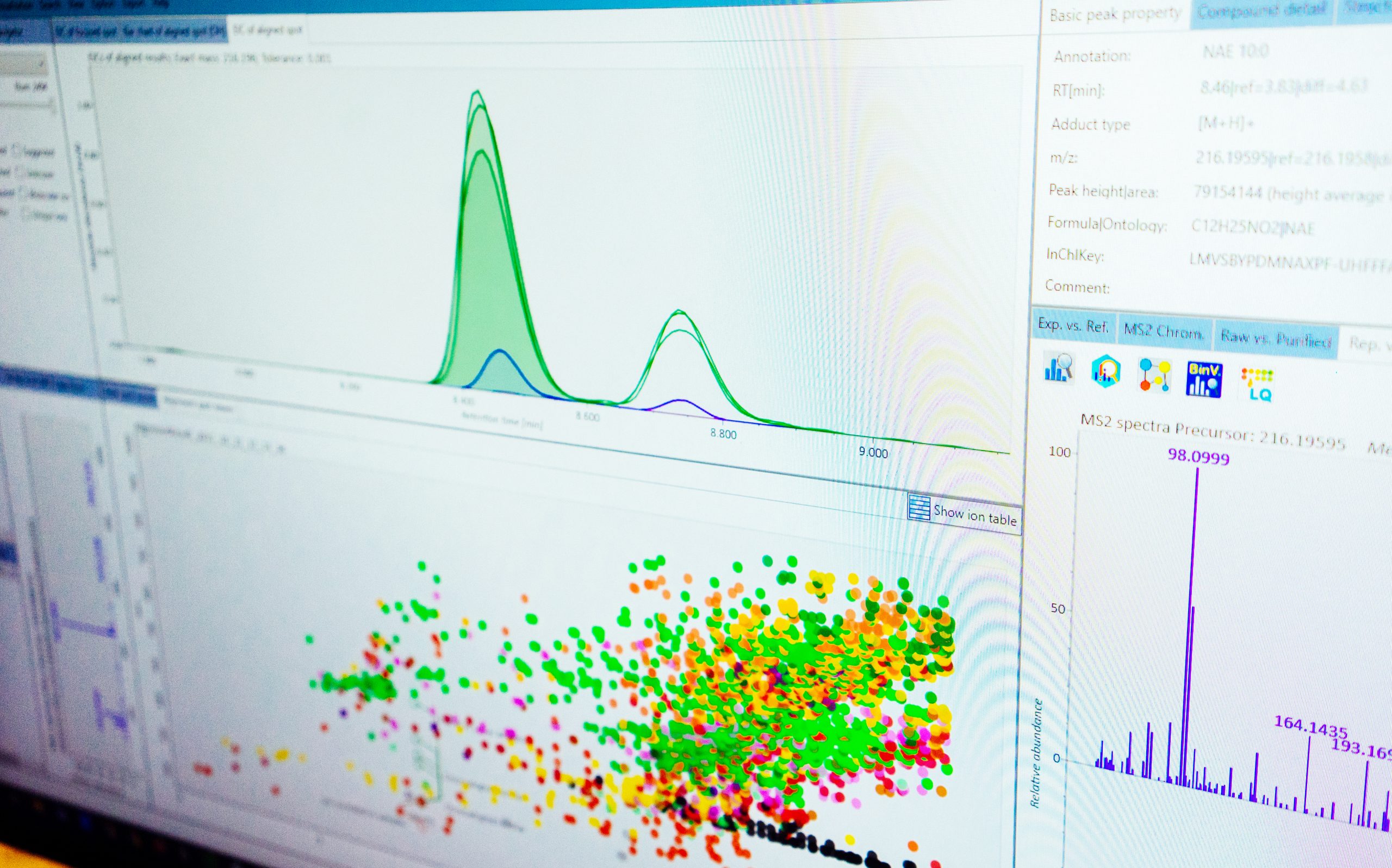
We offer coverage of the chemical exposome. Flexible services are based on the research study hypotheses and user needs, including sample preparation for different human biofluids and environmental samples, and combination of different mass spectrometry analyses.
- Comprehensive trace analysis by liquid (LC) or gas chromatography (GC) with high-resolution mass-spectrometry detection using Orbitrap technology.
- Combined target and nontarget acquisition by data-dependent (DDA) and data-independent (DIA) MS/MS, with electrospray or atmospheric pressure chemical ionization
- Sample preparation in ultratrace clean laboratory for a range of environmental and human samples (water, air, human plasma or urine, wildlife tissues)
- In-line solid phase extraction for water or biofluids
- Structural elucidation of unknown small molecules at trace concentration in complex samples
- Data processing with open science software and FAIR workflows
- Multivariate data analysis
Equipment
Positive pressure high efficiency particulate (HEPA)-filtered clean laboratory.
High-resolution mass spectrometers:
- UHPLC 480 Exploris Orbitrap, LC-HRMS
- UHPLC Q Exactive HF-X Orbitrap, LC-HRMS
- GC Q Exactive Orbitrap, GC-HRMS