CRISPR Functional Genomics
Don’t miss the open call for pipeline projects within the Chemical Biology and Genome Engineering Platform! Deadline October 11.
Unlock the power of CRISPR with CRISPR Functional Genomics (CFG)! We provide affordable precision gene-editing and massively parallel functional genomic screens using various CRISPR-systems.
Massively parallel perturbation in pooled CRISPR screens lets you investigate tens of thousands of genes or genetic elements for their involvement in the biological processes that matter to you. CFG provides verified lentiviral CRISPR guide libraries for whole-genome or targeted loss- and gain-of function studies, including CRISPR-KO, CRISPR-I, and CRISPR-A. To meet your unique research needs, we create any custom guide library – from design to packaged lentivirus.
Services span the entire pooled screening process, from Cas-cell line generation and library virus creation, via phenotypic selection by FACS or live/dead, to gDNA and NGS library preparation, NGS at NGI, and data analysis.
Together with Eukaryotic Single Cell Genomics (ESCG) we provide a pipeline for pooled screens with single cell RNASeq readout (Perturb-Seq, CROP-Seq).
We are constantly innovating and are developing base-editing approaches for mutagenesis screens to study protein-drug interactions, as well as optical pooled screens for complex phenotypic readouts. Accelerate your research with CFG’s state-of-the-art methods, or develop new CRISPR technologies together with us!
Our second area of expertise is precision edits in cell lines, such as knock-out, knock-in, deletions, point mutations, etc. We collborate with the Karolinska Center for Transgene Technologies (KCTT) to offer CRISPR technology in mouse, and with the Cell and Gene Therapy Core at Lund University for precision editing of iPSCs and ESCs.
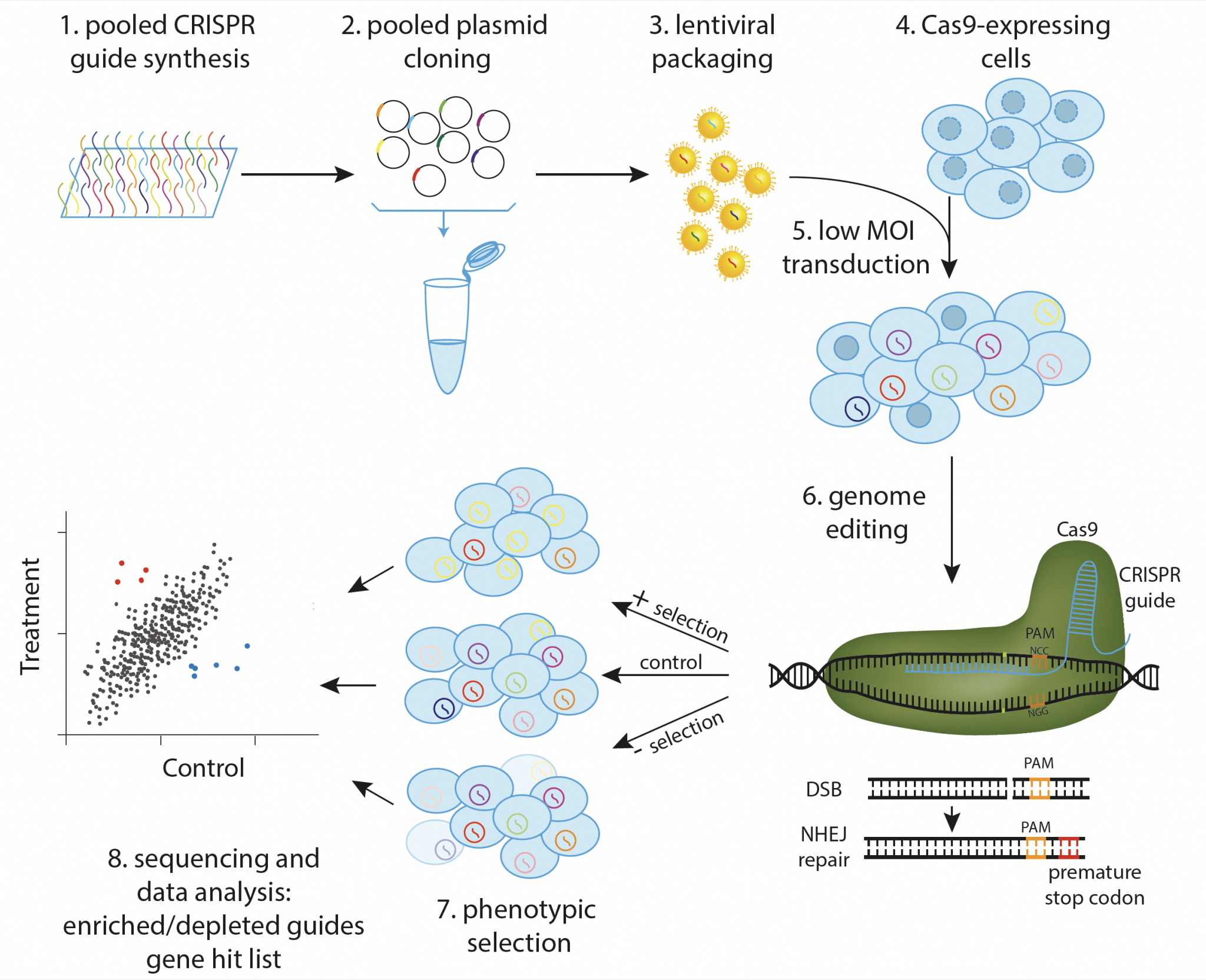
Standard pooled CRISPR screen. Guides targeting the elements of interest are synthesized, cloned, and packaged into lentivirus in pool (1-3). Cas9-expressing cells (4) are then transduced at low multiplicity of infection to ensure that each cell obtains a single guide (5), and phenotypic selection is applied after genome editing has occurred (6,7). Guide representation in treatment and control populations is then determined by NGS, resulting in a list of genes involved in the biological process of interest (8).
Barcoded guide libraries.
For superior statistics and data analysis, all our libraries are barcoded, as described in our publication CRISPR/Cas9 Screening Using Unique Molecular Identifiers. PDF. Full text.
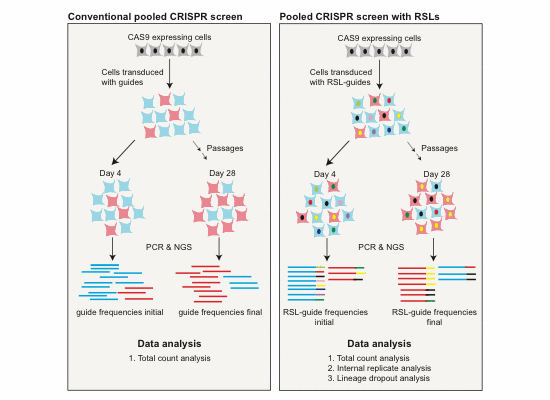
- Compared to the conventional method, inclusion of RSLs generates considerably more information at an identical experimental scale and enables additional ways of data analysis.
- RSL‐based analysis requires fewer cells per guide to reach a set statistical power. This is important if cell numbers are limiting, such as in very large, genome‐wide screens and/or screens in primary cells.
- RSLs can be used to track cell lineages throughout a pooled screen.
Services
- Cas-expressing cell line generation (Cas9, nCas9, dCas9, dCas9 fusions, Cas12a, Cas13d)
- CRISPR-ko, CRISPR-inhibition, CRISPR-activation screens
- Multiplexed Cas12a screens
- Base-editor screens
- RNA targeting screens with Cas13d
- Small pooled screens with single cell transcriptomic readout
- Experienced support for screen design
- Custom-made lentiviral guide libraries produced and provided
- Library transduction performed
- NGS libraries prepared
- NGS at NGI Stockholm included in the pipeline
- Data analysis: hit list generated
Applications
- Determine essential genes in specific cell types
- Screen for drug resistance/sensitivity genes
- Find novel genes/pathways involved in any cellular process
- Map novel genes/pathways regulating a reporter gene
Using the unit
CFG is open to the entire Swedish and international research community, academia, healthcare and industry alike. Please contact us to schedule a free consultation meeting.
Your project is out of the box? All the better, we would love to hear from you!
Shipping and Mailing Address
Biomedicum 9B
Tomtebodavägen 16
171 65 Solna
Visiting Address
Biomedicum 9B
Solnavägen 9
171 65 Solna