SciLifeLab Spatial Omics
Welcome to explore the spatial capabilities offered at SciLifeLab!
Here you will find an overview of spatial omics methods offered at SciLifeLab. More details on the different spatial technologies and services can be found following the links to the different units provided in the left section.



Spatially resolved omics technologies have emerged during the last years and have undoubtedly changed the way we understand the spatial organization of complex multicellular biological systems. SciLifeLab researchers have contributed deeply to the development of spatial omics methods (1, 2, 3) and many of these technologies as well as others are offered as a service within the SciLifeLab infrastructure. From 2021 the services within spatial omics are gathered within our newly formed platform of Spatial and Single Cell Biology. With this organisation we hope to better coordinate and support users who wish to apply several spatial omics methods in their research projects. The Spatial and Single Cell Biology platform covers cutting-edge technologies for spatial profiling of transcripts, proteins, DNA and small molecules, as well as several approaches for single cell analysis, which preferably can be combined within your spatial omics study.
1. Ståhl & Salmén et al. Science (2016), 2. Ke et al. Nature Methods (2013), 3. Thul et al, Science (2017)

If you are interested in finding out more about how we can support projects that can benefit from a spatial multiomics approach, please get in touch with our Platform Coordinator to discuss your next spatial project further:
Charlotte Stadler: charlotte.stadler@scilifelab.se
Or, contact the Head of Unit offering the technology you are interested in (contact information can be found in the left section)
Services
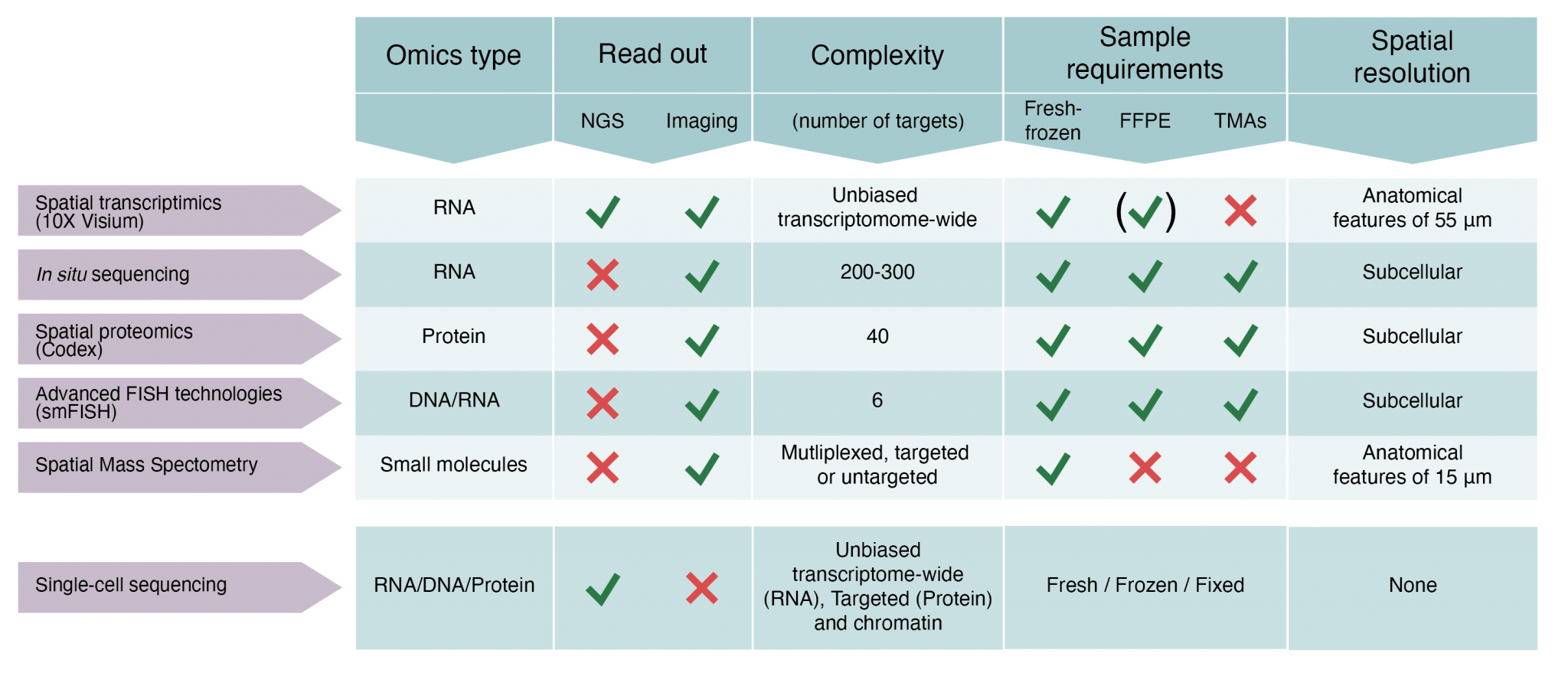
We offer access to technology for spatially resolved RNA, protein and small molecule analyses of histological tissue sections. The spatial capabilities established at SciLifeLab comprise expertise around five state-of-the-art techniques. Details about each technology are provided in Figure 1.
(i) Spatial transcriptomics (transcriptome-wide spatial gene expression analysis in tissue sections using the 10X Genomics Visium assay)
(ii) In Situ Sequencing (targeted gene expression profiling in tissue sections at subcellular resolution)
(iii) Spatial Proteomics (multiplexed (up to 40 plex) targeted spatial proteomics analysis at single cell level in tissue sections using the Phenocycler platform). Low plex high content immunofluorescence of cell cultures in 96 or 384 well format is also offered using the Human Protein Atlas antibody resource.
(iv) Advanced FISH technologies (smFISH) (targeted gene expression profiling (up to 6 targets) in cells or tissue sections at subcellular resolution)
(v) Spatial Mass Spectrometry (untargeted or targeted metabolomics analysis in tissue sections)
Sample Requirements
Spatial Transcriptomics 10X Visium
Fresh frozen OCT embedded tissue, with intact morphology and high RNA quality. We require our users to do their own QC steps before sending samples, more info found here. Please keep in mind that the maximum size of the tissue block has to be not more than 6×6 mm (not larger than the predefined capture area of the array). For FFPE tissue samples (human or mouse), please contact us for a discussion.
In Situ Sequencing
Fresh frozen or PFA fixed OCT embedded tissue sample. FFPE tissue or TMA samples are also suitable. 8-12 μm tissue sections are optimal. Collect on SuperFrost plus microscope slides, and store slides in -80°C until usage. The sections/accommodated sections should not be placed towards the edge of the slide (at least 4mm away from the edge), and the space in between the sections/spot should be at least 8mm. It is highly recommended to discuss the sample preparation with the Head of Unit in advance.
Spatial Proteomics
Highly multiplexed immunofluorescence with CODEX: Fresh frozen OCT embedded tissue sections (mouse ur human) or FFPE tissue sections (human). Sections should be < 10 μm thick and fit onto a 22×22 mm coverslip. Note that the data capture area is limited to 11×11 mm, centered onto the coverslip. Whole tissue sections or tissue microarrays (TMA) can be mounted on the coverslip. We urge users to contact the Head of Unit for receiving detailed instructions on sample preparation and shipment. For high throughput standard immunofluorescence of cell lines and primary cell cultures in 96 or 384 well plates, contact the Head of Unit for fixation and pre-staining sample preparation protocols.
Advanced FISH technologies
For smFISH: cells attached onto microscope coverslips fixed with 4% PFA/1xPBS (10 min at RT) followed by 70% EtOH, or methanol-acetic acid 3:1; frozen OCT tissue sections (~10 μm) mounted on microscope coverslips pre-coated with poly-L-lysine and postfixed with 4% PFA/1xPBS (10 min at RT) or methanol-acetic acid 3:1; FFPE tissue sections (5-10 μm) cut in RNase-free conditions, mounted on IHC-grade microscope slides, and shipped dry. We urge users to contact the Head of Unit for detailed instructions on sample preparation and shipment.
For DNA FISH: cells attached onto microscope coverslips fixed with 4% PFA/1xPBS (10 min at RT); FFPE tissue sections/TMAs (5-10 μm) mounted on IHC-grade microscope slides and shipped dry. We urge users to contact the Head of Unit for detailed instructions on sample preparation and shipment.
Spatial Mass Spectrometry
Fresh frozen, non-embedded tissue samples should be sectioned, typically at 10-14 µm, using a cryomicrotome. For matrix-assisted laser desorption ionization (MALDI) mass spectrometry imaging (MSI), tissue sections need to be mounted on indium thin oxide coated glass slides. Regular glass slides should be used for desorption electrospray ionization (DESI) MSI. Fragile or small tissue samples can be embedded in mass spectrometry compatible media, e.g., gelatin. Please contact us for more information about embedding protocols.
What We Deliver
Spatial Transcriptomics 10X Visium
Sequencing data QC is handled by the National Genomics Infrastructure and run through the 10X Genomics Space Ranger analysis pipeline. Raw data, jpeg images and Space Ranger output files are delivered. Read more about the Spatial Transcriptomics analysis pipeline here.
In Situ Sequencing
Post processed images (projected and stitched images as TIFF) and x-y coordinates of all spots/signals in csv file format. RAW image delivery is also available as an option. A visualization tool for the x-y coordinates of all spots will also be provided, e.g. mapping all signals on top of the DAPI image.
Spatial Proteomics
CODEX data: The unit delivers raw data images and post processed images (including deconvolution, illumination correction, stitching and cell segmentation). An fcs file with quantitative measurements of all markers for every cell is delivered, allowing for downstream analysis using open source software. A technical quality control and report with detailed assay information for manuscript preparation is delivered.
Advanced FISH technologies
We deliver the following products/data:
1) Design of DNA/RNA FISH oligonucleotide probes against a species of interest (provided the availability of a good genome/transcriptome reference)
2) DNA/RNA FISH oligonucleotide probes, either ready-to-use or as PCR products for downstream production of ssDNA probes (for services involving custom probe production)
3) Raw DNA/RNA FISH images (nd2 and tiff files) (for services involving DNA and/or RNA FISH on samples provided by the Customer). Optional: deconvolved image files; cell segmentation masks based on DNA staining.
Spatial Mass Spectrometry
Spatial mass spectrometry is a sensitive molecular imaging technique that provides combined molecular information and spatial resolution. It allows localization, identification and quantification of drugs, their metabolites and endogenous biomolecules such as neurotransmitters, lipids, peptides, and small proteins directly in tissue sections at near-cellular lateral resolution. The technique is label-free and multiplex allowing simultaneous detection of hundreds to thousands of molecules based on their molecular mass. Depending on the project’s needs, the MSI analyses could be performed targeted or untargeted. Quantitative measurements can be performed for specific target molecules.
Data Analysis Support
While the spatial omics units are experts in data generation using the technologies described, our support in analyzing the data is limited to initial primary analysis. If your group lacks bioinformatic competence, we highly recommend you to contact NBIS for further support on downstream analysis, visualization, and integration of spatial datasets at an early stage of the project. This contact can also be provided by the Head of Unit of the relevant spatial omics method upon project discussion.
Examples Of Successful User Projects
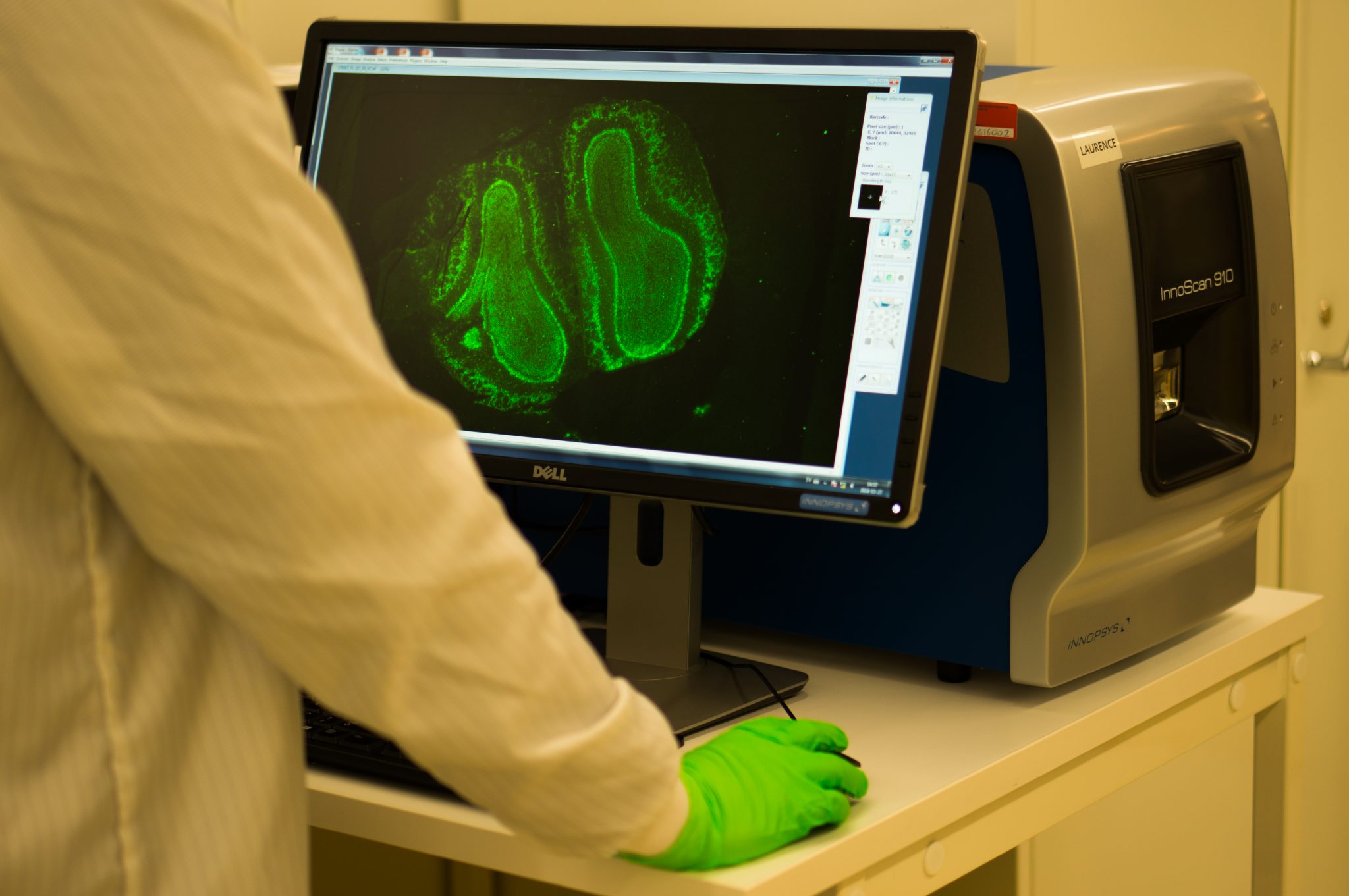
A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart
A spatial multi omics project which combined spatial transcriptomics, in situ sequencing, spatial proteomics and single-cell RNA-sequencing to create a spatial cellular map of human cardiogenesis.
"The process of cardiac morphogenesis in humans is incompletely understood. Its full characterization requires a deep exploration of the organ-wide orchestration of gene expression with a single-cell spatial resolution. Here, we present a molecular approach that reveals the comprehensive transcriptional landscape of cell types populating the embryonic heart at three developmental stages and that maps cell-type-specific gene expression to specific anatomical domains. Spatial transcriptomics identified unique gene profiles that correspond to distinct anatomical regions in each developmental stage. Human embryonic cardiac cell types identified by single-cell RNA sequencing confirmed and enriched the spatial annotation of embryonic cardiac gene expression. In situ sequencing was then used to refine these results and create a spatial subcellular map for the three developmental phases. Finally, we generated a publicly available web resource of the human developing heart to facilitate future studies on human cardiogenesis.“
Read the full article here.